4XBG
 
 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA): 2.73 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-16 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
3EQI
 
 | |
5RF1
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with NCL-00023830 | | Descriptor: | 3C-like proteinase, 4-bromobenzene-1-sulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z979145504 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl (2-methyl-4-phenyl-1,3-thiazol-5-yl)carbamate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
1UY4
 
 | |
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5B4C
 
 | | Crystal structure of H10N mutant of LpxH with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
2WNH
 
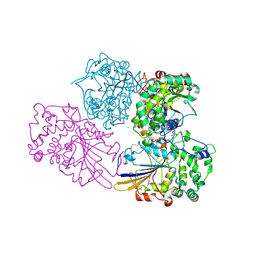 | | Crystal Structure Analysis of Klebsiella sp ASR1 Phytase | | Descriptor: | 3-PHYTASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bohm, K, Mueller, J.J, Heinemann, U. | | Deposit date: | 2009-07-09 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Klebsiella Sp. Asr1 Phytase Suggests Substrate Binding to a Preformed Active Site that Meets the Requirements of a Plant Rhizosphere Enzyme.
FEBS J., 277, 2010
|
|
5RL9
 
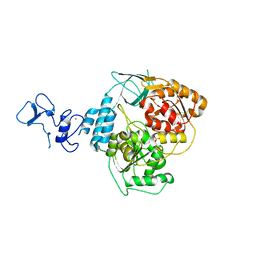 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1703168683 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)methanesulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RHS
 
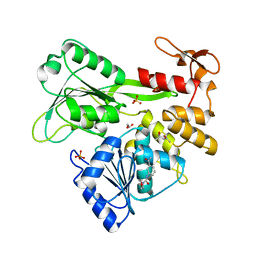 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z274555794 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, N-(3-acetylphenyl)morpholine-4-carboxamide, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RS7
 
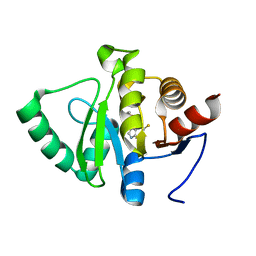 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000034618676 | | Descriptor: | 1-{2-[(propan-2-yl)oxy]ethyl}-2-sulfanylidene-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5O3F
 
 | | Human Brd2(BD2) mutant in complex with ET-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-butanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Runcie, A.C, Chan, K.-H, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
7DGB
 
 | |
7DGF
 
 | |
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
3EO9
 
 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7DGG
 
 | |
7DHJ
 
 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
7DG6
 
 | |
7HIE
 
 | | Group deposition for combi-soaks of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1041785508 and Z905065822 (CHIKV_MacB-x1488) | | Descriptor: | (5R)-1,3,7-triazaspiro[4.5]decane-2,4-dione, (5S)-1,3,7-triazaspiro[4.5]decane-2,4-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Group deposition for combi-soaks of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
4PTC
 
 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1UXX
 
 | | CBM6ct from Clostridium thermocellum in complex with xylopentaose | | Descriptor: | CALCIUM ION, XYLANASE U, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Henrissat, D.B.B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
7HIQ
 
 | | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with RA-0188437-02 (CHIKV_MacB-x2350) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Almahli, H, Balcomb, B.H, Capkin, E, Chandran, A.V, Chen, W, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Saleem, R.S.Z, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Todd, M.H, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Group deposition of Chikungunya virus nsP3 macrodomain in complex with inhibitors from the READDI-AC AViDD center
To Be Published
|
|
7HIG
 
 | | Group deposition for combi-soaks of Chikungunya virus nsP3 macrodomain -- Crystal structure of Chikungunya virus nsP3 macrodomain in complex with Z1041785508 and Z1165350851 (CHIKV_MacB-x1495) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-hydroxyethyl)-5-methyl-1,2-dihydro-3H-pyrazol-3-one, 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, ... | | Authors: | Aschenbrenner, J.C, Fairhead, M, Godoy, A.S, Balcomb, B.H, Capkin, E, Chandran, A.V, Golding, M, Koekemoer, L, Lithgo, R.M, Marples, P.G, Ni, X, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Xavier, M.-A.E, Fearon, D, von Delft, F. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Group deposition for combi-soaks of Chikungunya virus nsP3 macrodomain
To Be Published
|
|
3EQF
 
 | |
