7LWT
 
 | |
3H0A
 
 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3QHM
 
 | | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | Descriptor: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
4J3W
 
 | | Crystal structure of barley limit dextrinase (E510A mutant) in complex with a branched maltohexasaccharide | | Descriptor: | CALCIUM ION, IODIDE ION, Limit dextrinase, ... | | Authors: | Sim, L, Windahl, M.S, Moeller, M.S, Henriksen, A. | | Deposit date: | 2013-02-06 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase
J.Mol.Biol., 427, 2015
|
|
3CK9
 
 | | B. thetaiotaomicron SusD with maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SusD, ... | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
3HGP
 
 | | Structure of porcine pancreatic elastase complexed with a potent peptidyl inhibitor FR130180 determined by high resolution crystallography | | Descriptor: | 4-[[(2S)-3-methyl-1-oxo-1-[(2S)-2-[[(3S)-1,1,1-trifluoro-4-methyl-2-oxo-pentan-3-yl]carbamoyl]pyrrolidin-1-yl]butan-2-yl]carbamoyl]benzoic acid, CALCIUM ION, Elastase-1, ... | | Authors: | Tamada, T, Kinoshita, T, Kuroki, R, Tada, T. | | Deposit date: | 2009-05-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Combined High-Resolution Neutron and X-ray Analysis of Inhibited Elastase Confirms the Active-Site Oxyanion Hole but Rules against a Low-Barrier Hydrogen Bond
J.Am.Chem.Soc., 131, 2009
|
|
3A5V
 
 | | Crystal structure of alpha-galactosidase I from Mortierella vinacea | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kobayashi, H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tetramer Structure of the Glycoside Hydrolase Family 27 alpha-Galactosidase I from Umbelopsis vinacea
Biosci.Biotechnol.Biochem., 73, 2009
|
|
1J88
 
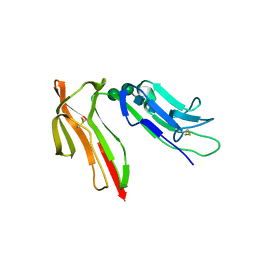 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), TETRAGONAL CRYSTAL FORM 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
3D7H
 
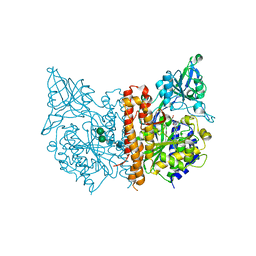 | |
4B71
 
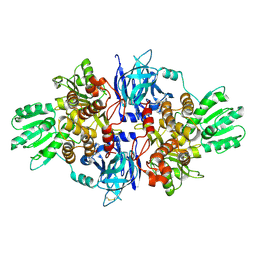 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propan-1-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
3UAN
 
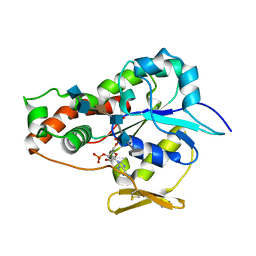 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7DJR
 
 | | Crystal structure of SARS-CoV-2 main protease (no ligand) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Deetanya, P, Wangkanont, K. | | Deposit date: | 2020-11-21 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification.
Comput Struct Biotechnol J, 19, 2021
|
|
5R9I
 
 | |
1MAA
 
 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
7DKS
 
 | |
3EQD
 
 | |
5WZE
 
 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
5ZCB
 
 | | Crystal structure of Alpha-glucosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Saburi, W, Yao, M. | | Deposit date: | 2018-02-16 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Function and structure of GH13_31 alpha-glucosidase with high alpha-(1→4)-glucosidic linkage specificity and transglucosylation activity.
FEBS Lett., 592, 2018
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
3HG1
 
 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
3HD6
 
 | | Crystal Structure of the Human Rhesus Glycoprotein RhCG | | Descriptor: | Ammonium transporter Rh type C, octyl beta-D-glucopyranoside | | Authors: | Gruswitz, F, Chaudhary, S, Ho, J.D, Pezeshki, B, Ho, C.-M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-05-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of human Rh based on structure of RhCG at 2.1 A.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3E6P
 
 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
5WZM
 
 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
