1Y5I
 
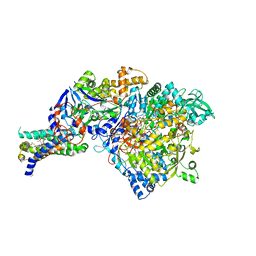 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1XVE
 
 | |
1Y4Z
 
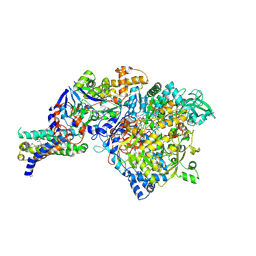 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5N
 
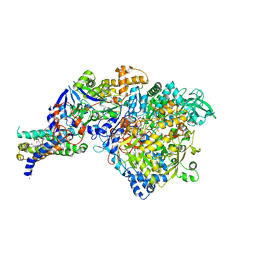 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5L
 
 | | The crystal structure of the NarGHI mutant NarI-H66Y | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Z5X
 
 | | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor ligand binding domain, PHOSPHATE ION, ... | | Authors: | Carmichael, J.A, Lawrence, M.C, Graham, L.D, Pilling, P.A, Epa, V.C, Noyce, L, Lovrecz, G, Winkler, D.A, Pawlak-Skrzecz, A. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design.
J.Biol.Chem., 280, 2005
|
|
1ZKY
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-3M and A Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide | | Descriptor: | 4-[(1S,2S,5S)-5-(HYDROXYMETHYL)-6,8,9-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Hahm, J.B, Nettles, K.W, Greene, G.L. | | Deposit date: | 2005-05-04 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
1ZS5
 
 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
3ZSX
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-({[2-(benzylcarbamoyl)benzyl](prop-2-en-1-yl)amino}methyl)-1,3-benzodioxole-4-carboxylate, ACETATE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
4AFE
 
 | | Nek2 bound to hybrid compound 21 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-AMINO-5-{4-[(DIMETHYLAMINO)METHYL]THIOPHEN-2-YL}PYRIDIN-3-YL)-2-{[(1R,2Z)-4,4,4-TRIFLUORO-1-METHYLBUT-2-EN-1-YL]OXY}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Yeoh, S, Innocenti, P, Hoelder, S, Bayliss, R. | | Deposit date: | 2012-01-19 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Design of Potent and Selective Hybrid Inhibitors of the Mitotic Kinase Nek2: Structure-Activity Relationship, Structural Biology, and Cellular Activity.
J.Med.Chem., 55, 2012
|
|
6GJB
 
 | | Erk2 signalling protein | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, [(1~{R},4~{Z})-cyclooct-4-en-1-yl] ~{N}-[4-[4-[[4-[1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-2-oxidanylidene-pyridin-4-yl]pyrimidin-2-yl]amino]pyridin-2-yl]but-3-ynyl]carbamate | | Authors: | O'Reilly, M. | | Deposit date: | 2018-05-16 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe.
Chem Sci, 9, 2018
|
|
6H4H
 
 | | Usp28 catalytic domain variant E593D in complex with UbPA | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28, ... | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H7P
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH4, cyclohexanone and allyl amine | | Descriptor: | CYCLOHEXANONE, Reductive Aminase, [[(2~{R},3~{S},4~{R},5~{R})-5-[(3~{R})-3-aminocarbonylpiperidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, ... | | Authors: | Sharma, M, Grogan, G, Mangas-Sanchez, J, Turner, N.J. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of Reductive Aminase from Aspergillus terreus
Acs Catalysis, 2018
|
|
6HQB
 
 | | Monomeric cyanobacterial photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Netzer-El, S.Y, Nelson, N, Caspy, I. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-16 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of Photosystem I Monomer From Synechocystis PCC 6803.
Front Plant Sci, 9, 2018
|
|
6GZS
 
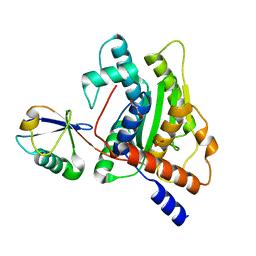 | |
4AJ4
 
 | | rat LDHA in complex with 4-((2-allylsulfanyl-1,3-benzothizol-6-yl) amino)-4-oxo-butanoic acid | | Descriptor: | 4-oxo-4-{[2-(prop-2-en-1-ylsulfanyl)-1,3-benzothiazol-6-yl]amino}butanoic acid, L-LACTATE DEHYDROGENASE A CHAIN, MALONATE ION | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
6HUG
 
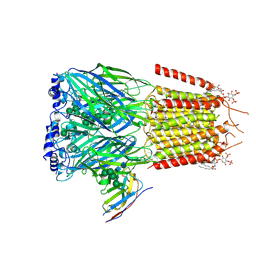 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with picrotoxin and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
3ZOH
 
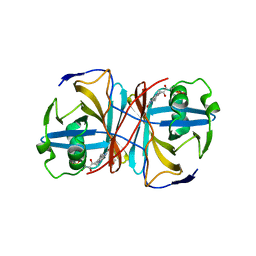 | | Crystal structure of FMN-binding protein (YP_005476) from Thermus thermophilus with bound 1-Cyclohex-2-enone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOREDOXIN, cyclohex-2-en-1-one | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, C.C, Steiner, K, Winkler, C, Schwamberger, O, Schwab, H, Faber, K, Gruber, K. | | Deposit date: | 2013-02-21 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of Promiscuous Ene-Reductase Activity by Mining Structural Databases Using Active Site Constellations.
Nat.Commun., 5, 2014
|
|
3ZK5
 
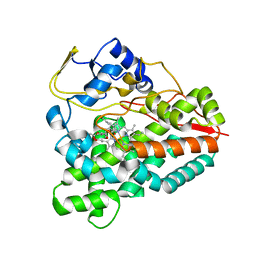 | | PikC D50N mutant bound to the 10-DML analog with the 3-(N,N-dimethylamino)ethanoate anchoring group | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ethyl-3,5,7,11-tetramethyl-2,8-dioxooxacyclododec-9-en-4-yl N,N-dimethylglycinate, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M. | | Deposit date: | 2013-01-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Directing Group-Controlled Regioselectivity in an Enzymatic C-H Bond Oxygenation.
J.Am.Chem.Soc., 136, 2014
|
|
6HOP
 
 | |
6HUJ
 
 | | CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with picrotoxin, GABA and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
3ZSO
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based design | | Descriptor: | 5-{[(2-{[bis(4-methoxyphenyl)methyl]carbamoyl}benzyl)(prop-2-en-1-yl)amino]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, INTEGRASE, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Deadman, J.J, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
6H45
 
 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
3ZVU
 
 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
3ZSR
 
 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-06-30 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
