1VL7
 
 | |
5YPM
 
 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem representing an EI1 complex | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, Metallo-beta-lactamase NDM-1, SULFATE ION, ... | | Authors: | Feng, H, Wang, D, Liu, W. | | Deposit date: | 2017-11-02 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis.
Nat Commun, 8, 2017
|
|
3UPK
 
 | | E. cloacae MURA in complex with UNAG | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4HMB
 
 | | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution | | Descriptor: | 3-{3-[(DIMETHYLAMINO)METHYL]-1H-INDOL-7-YL}PROPAN-1-OL, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Shukla, P.K, Haridas, M, Chandra, D.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-18 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the complex of group II phospholipase A2 with a 3-{3-[(Dimethylamino)methyl]-1H-indol-7-yl}propan-1-ol at 2.21 A Resolution
To be Published
|
|
3BC4
 
 | | I84V HIV-1 protease in complex with a pyrrolidine diester | | Descriptor: | 2-aminoethyl naphthalen-1-ylacetate, DIMETHYL SULFOXIDE, protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-11-12 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Targeting the open-flap conformation of HIV-1 protease with pyrrolidine-based inhibitors
Chemmedchem, 3, 2008
|
|
5VEA
 
 | |
1IT6
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | Descriptor: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | Authors: | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
6VI0
 
 | | Cryo-EM structure of VRC01.23 in complex with HIV-1 Env BG505 DS.SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A matrix of structure-based designs yields improved VRC01-class antibodies for HIV-1 therapy and prevention.
Mabs, 13
|
|
2WIY
 
 | | Cytochrome P450 XplA heme domain P21212 | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450-LIKE PROTEIN XPLA, IMIDAZOLE, ... | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
3KC0
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 10b | | Descriptor: | Fructose-1,6-bisphosphatase 1, [(8H-indeno[1,2-d][1,3]thiazol-4-yloxy)methyl]phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1PYE
 
 | | Crystal structure of CDK2 with inhibitor | | Descriptor: | Cell division protein kinase 2, [2-AMINO-6-(2,6-DIFLUORO-BENZOYL)-IMIDAZO[1,2-A]PYRIDIN-3-YL]-PHENYL-METHANONE | | Authors: | Zhang, F, Hamdouchi, C. | | Deposit date: | 2003-07-08 | | Release date: | 2004-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo[1,2-a]pyridines.
MOL.CANCER THER., 3, 2004
|
|
4KXV
 
 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
1LB3
 
 | | Structure of recombinant mouse L chain ferritin at 1.2 A resolution | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, GLYCEROL, ... | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Chevalier, J.-M, Precigoux, G, Santambrogio, P, Arosio, P. | | Deposit date: | 2002-04-02 | | Release date: | 2003-01-28 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural description of the active sites of mouse L-chain ferritin at 1.2A resolution
J.Biol.Inorg.Chem., 8, 2003
|
|
5EOE
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 (orthorhombic form) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3K5F
 
 | | Human BACE-1 COMPLEX WITH AYH011 | | Descriptor: | (1R,3S)-3-[1-(acetylamino)-1-methylethyl]-N-[(1S,2S,4R)-1-benzyl-5-(butylamino)-2-hydroxy-4-methyl-5-oxopentyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
7JN1
 
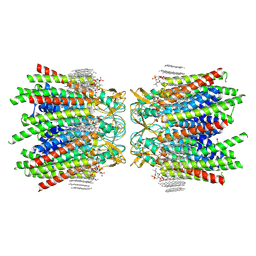 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
3BYO
 
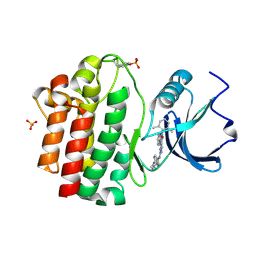 | | X-Ray co-crystal structure of 2-amino-6-phenylpyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one 25 bound to Lck | | Descriptor: | 6-(2,6-dimethylphenyl)-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-one, Proto-oncogene tyrosine-protein kinase LCK, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel 2-amino-6-phenyl-pyrimido[5',4':5,6]pyrimido[1,2-a]benzimidazol-5(6H)-ones as potent and orally active inhibitors of lymphocyte specific kinase (Lck): synthesis, SAR, and in vivo anti-inflammatory activity.
J.Med.Chem., 51, 2008
|
|
3KBZ
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 6 | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(2-amino-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7UX3
 
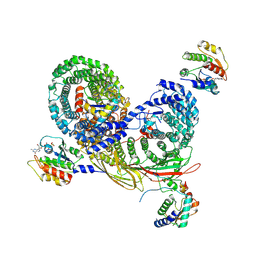 | | Asymmetric unit of AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-05-04 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
3KC1
 
 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor 19a | | Descriptor: | Fructose-1,6-bisphosphatase 1, {[(7-carbamoyl-8H-indeno[1,2-d][1,3]thiazol-4-yl)oxy]methyl}phosphonic acid | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-10-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based drug design of tricyclic 8H-indeno[1,2-d][1,3]thiazoles as potent FBPase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1NU2
 
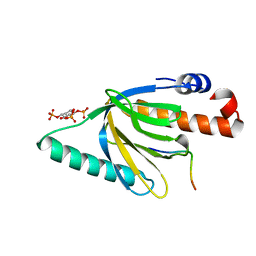 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NWG
 
 | | BETA-1,4-GALACTOSYLTRANSFERASE COMPLEX WITH ALPHA-LACTALBUMIN AND N-BUTANOYL-GLUCOAMINE | | Descriptor: | 2-(butanoylamino)-2-deoxy-beta-D-glucopyranose, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Shah, P.S, Qasba, P.K. | | Deposit date: | 2003-02-06 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | ALPHA-LACTALBUMIN (LA) STIMULATES MILK
BETA-1,4-GALACTOSYLTRANSFERASE I (BETA 4GAL-T1) TO
TRANSFER GLUCOSE FROM UDP-GLUCOSE TO
N-ACETYLGLUCOSAMINE. CRYSTAL STRUCTURE OF BETA
4GAL-T1 X LA COMPLEX WITH UDP-GLC.
J.Biol.Chem., 276, 2001
|
|
