4BKX
 
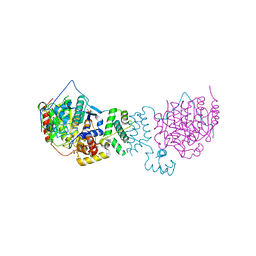 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
9HCT
 
 | | Crystal structure of human TRF1 TRFH domain in complex with compound 50 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide, CALCIUM ION, ... | | Authors: | Casale, G, Le Bihan, Y.-V, van Montfort, R.L.M, Guettler, S. | | Deposit date: | 2024-11-11 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of first-in-class inhibitors of the TRF1-TIN2 protein-protein interaction by fragment screening
To Be Published
|
|
4PCE
 
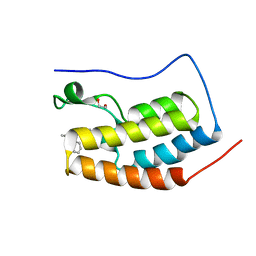 | | Crystal Structure of the first bromodomain of human BRD4 in complex with compound B13 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-2-ethyl-1,5,6,7-tetrahydro-4H-indol-4-one, Bromodomain-containing protein 4 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5X9O
 
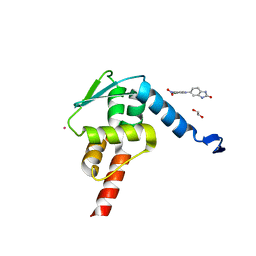 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
4B1F
 
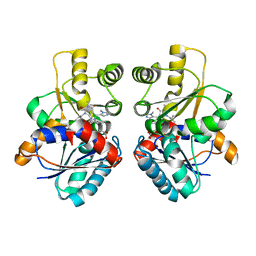 | | Design of Inhibitors of Helicobacter pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty | | Descriptor: | 5-methyl-3-(1-methyl-1H-imidazol-5-yl)-7-(2-methylpropyl)-2-(naphthalen-1-ylmethyl)-2H-pyrazolo[3,4-d]pyrimidine-4,6(5H,7H)-dione, D-GLUTAMIC ACID, GLUTAMATE RACEMASE | | Authors: | Basarab, G.S, Hill, P, Eyermann, C.J, Gowravaram, M, Kack, H, Osimoni, E. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design of Inhibitors of Helicobacter Pylori Glutamate Racemase as Selective Antibacterial Agents: Incorporation of Imidazoles Onto a Core Pyrazolopyrimidinedione Scaffold to Improve Bioavailabilty.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
9HD1
 
 | | Crystal structure of human TRF1 TRFH domain in complex with compound 59 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-propan-2-ylurea, ... | | Authors: | Casale, G, Le Bihan, Y.-V, van Montfort, R.L.M, Guettler, S. | | Deposit date: | 2024-11-11 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of first-in-class inhibitors of the TRF1-TIN2 protein-protein interaction by fragment screening
To Be Published
|
|
6JYQ
 
 | | Crystal structure of uPA_H99Y in complex with 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide | | Descriptor: | 3-azanyl-5-(azepan-1-yl)-N-carbamimidoyl-6-(furan-2-yl)pyrazine-2-carboxamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Buckley, B, Jiang, L.G, Huang, M.D, Kelso, M, Ranson, M. | | Deposit date: | 2019-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | H99Y-6F-HMA-pH7
To Be Published
|
|
9HDA
 
 | | Crystal structure of human TRF1 TRFH domain in complex with compound 58 | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, Telomeric repeat-binding factor 1 | | Authors: | Casale, G, Le Bihan, Y.-V, Van Montfort, R.L.M, Guettler, S. | | Deposit date: | 2024-11-12 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of first-in-class inhibitors of the TRF1-TIN2 protein-protein interaction by fragment screening
To Be Published
|
|
3HL8
 
 | | Crystal structure of exonuclease I in complex with inhibitor BCBP | | Descriptor: | (5R)-3-tert-butyl-1-(6-chloro-1,3-benzothiazol-2-yl)-4,5-dihydro-1H-pyrazol-5-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5X8U
 
 | | Crystal Structure of the wild Human ROR gamma Ligand Binding Domain. | | Descriptor: | Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Noguchi, M, Nomura, A, Murase, K, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ternary complex of human ROR gamma ligand-binding domain, inverse agonist and SMRT peptide shows a unique mechanism of corepressor recruitment
Genes Cells, 22, 2017
|
|
4AW6
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
5B27
 
 | | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid | | Descriptor: | Fatty acid-binding protein, heart, PALMITIC ACID, ... | | Authors: | Matsuoka, D, Sugiyama, S, Kakinouchi, K, Niiyama, M, Murata, M, Matsuoka, S. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid.
To Be Published
|
|
5RWG
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z17497990 | | Descriptor: | 2-(2,4-dimethylphenoxy)-1-morpholin-4-yl-ethanone, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
8RD3
 
 | |
2X3X
 
 | | structure of mouse syndapin I (crystal form 1) | | Descriptor: | PROTEIN KINASE C AND CASEIN KINASE SUBSTRATE IN NEURONS PROTEIN 1 | | Authors: | Ma, Q, Rao, Y, Vahedi-Faridi, A, Saenger, W, Haucke, V. | | Deposit date: | 2010-01-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Molecular Basis for SH3 Domain Regulation of F-Bar-Mediated Membrane Deformation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HEX
 
 | | Cyclic residues in alpha/beta-peptide helix bundles: GCN4-pLI side chain sequence on an (alpha-alpha-beta) backbone with cyclic beta-residues at positions 1, 4, 19 and 28 | | Descriptor: | alpha/beta-peptide based on the GCN4-pLI side chain sequence with an (alpha-alpha-beta) backbone and cyclic beta-residues at positions 1, 4, 19 and 28 | | Authors: | Horne, W.S, Price, J.L, Gellman, S.H. | | Deposit date: | 2009-05-10 | | Release date: | 2010-04-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural consequences of beta-amino acid preorganization in a self-assembling alpha/beta-peptide: fundamental studies of foldameric helix bundles.
J.Am.Chem.Soc., 132, 2010
|
|
5RXJ
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1722836661 | | Descriptor: | 2-(1H-imidazol-1-yl)ethyl dimethylcarbamate, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
3HLM
 
 | | Crystal Structure of Mouse Mitochondrial Aspartate Aminotransferase/Kynurenine Aminotransferase IV | | Descriptor: | Aspartate aminotransferase, mitochondrial, GLYCEROL | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2009-05-27 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
Cell.Mol.Life Sci., 67, 2010
|
|
5RYE
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z33545544 | | Descriptor: | DIMETHYL SULFOXIDE, N~1~-phenylpiperidine-1,4-dicarboxamide, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
5RYQ
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z57258487 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ2
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z68277692 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
4MAQ
 
 | |
5R2A
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H02, DMSO-free | | Descriptor: | 3-cyclopentyl-1-(piperazin-1-yl)propan-1-one, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.049 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6KAB
 
 | |
1J87
 
 | | HUMAN HIGH AFFINITY FC RECEPTOR FC(EPSILON)RI(ALPHA), HEXAGONAL CRYSTAL FORM 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Sechi, S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2001-05-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms.
J.Mol.Biol., 311, 2001
|
|
