5M0H
 
 | |
1DZR
 
 | | RmlC from Salmonella typhimurium | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, GLYCEROL, SULFATE ION | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
5M0K
 
 | |
1E2F
 
 | | Human thymidylate kinase complexed with thymidine monophosphate, adenosine diphosphate and a magnesium-ion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ostermann, N, Schlichting, I, Brundiers, R, Konrad, M, Reinstein, J, Veit, T, Goody, R.S, Lavie, A. | | Deposit date: | 2000-05-22 | | Release date: | 2001-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights Into the Phosphoryltransfer Mechanism of Human Thymidylate Kinase Gained from Crystal Structures of Enzyme Complexes Along the Reaction Coordinate
Structure, 8, 2000
|
|
1DZ8
 
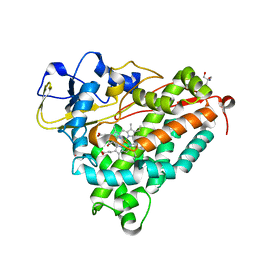 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
5M0N
 
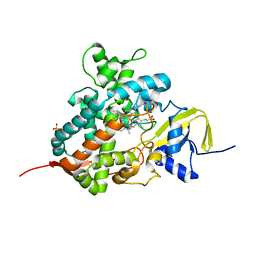 | | Crystal structure of cytochrome P450 OleT in complex with formate | | Descriptor: | FORMIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
1E1N
 
 | |
1E0N
 
 | |
1DWB
 
 | |
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5M0Z
 
 | |
1DBA
 
 | |
5M14
 
 | | Synthetic nanobody in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein, synthetic Nanobody L2_G11 (a-MBP#2) | | Authors: | Zimmermann, I, Egloff, P, Seeger, M.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Synthetic single domain antibodies for the conformational trapping of membrane proteins.
Elife, 7, 2018
|
|
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
5M1D
 
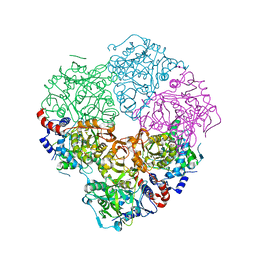 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5M56
 
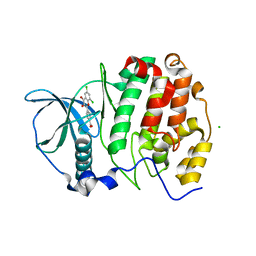 | | Monoclinic complex structure of human protein kinase CK2 catalytic subunit (isoform CK2alpha') with the inhibitor 4'-carboxy-6,8-chloro-flavonol (FLC21) | | Descriptor: | 4-[6,8-bis(chloranyl)-3-oxidanyl-4-oxidanylidene-chromen-2-yl]benzoic acid, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Niefind, K, Bischoff, N, Yarmoluk, S.M, Bdzhola, V.G, Golub, A.G, Balanda, A.O, Prykhod'ko, A.O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.237 Å) | | Cite: | Structural Hypervariability of the Two Human Protein Kinase CK2 Catalytic Subunit Paralogs Revealed by Complex Structures with a Flavonol- and a Thieno[2,3-d]pyrimidine-Based Inhibitor.
Pharmaceuticals, 10, 2017
|
|
1E0L
 
 | |
5M5J
 
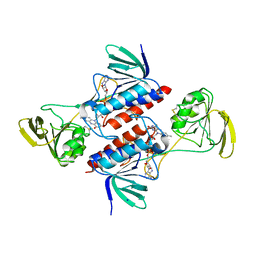 | | Thioredoxin reductase from Giardia duodenalis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, Thioredoxin reductase | | Authors: | Fiorillo, A, Ilari, A, Lalle, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural characterization of Giardia duodenalis thioredoxin reductase (gTrxR) and computational analysis of its interaction with NBDHEX.
Eur J Med Chem, 135, 2017
|
|
1E0M
 
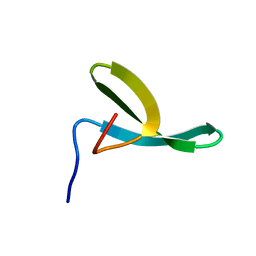 | |
5M5T
 
 | |
1DZD
 
 | | High resolution structure of acidic fibroblast growth factor (27-154), 24 NMR structures | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Lozano, R.M, Pineda-Lucena, A, Gonzalez, C, Jimenez, M.A, Cuevas, P, Redondo-Horcajo, M, Sanz, J.M, Rico, M, Gimenez-Gallego, G. | | Deposit date: | 2000-02-24 | | Release date: | 2000-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H-NMR Structural Characterization of a Non Mitogenic, Vasodilatory, Ischemia-Protector and Neuromodulatory Acidic Fibroblast Growth Factor
Biochemistry, 39, 2000
|
|
5M1L
 
 | |
1E41
 
 | |
1MMY
 
 | | Crystal structure of galactose mutarotase from Lactococcus lactis complexed with D-quinovose | | Descriptor: | Aldose 1-epimerase, SODIUM ION, alpha-D-quinovopyranose | | Authors: | Thoden, J.B, Kim, J, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-09-04 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and kinetic studies of sugar binding to galactose mutarotase from Lactococcus lactis.
J.Biol.Chem., 277, 2002
|
|
1DFB
 
 | | STRUCTURE OF A HUMAN MONOCLONAL ANTIBODY FAB FRAGMENT AGAINST GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE I | | Descriptor: | IGG1-KAPPA 3D6 FAB (HEAVY CHAIN), IGG1-KAPPA 3D6 FAB (LIGHT CHAIN) | | Authors: | He, X.M, Rueker, F, Casale, E, Carter, D.C. | | Deposit date: | 1992-03-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human monoclonal antibody Fab fragment against gp41 of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
