5S9T
 
 | |
5S9Z
 
 | |
5S9Y
 
 | |
1S5T
 
 | |
5SMJ
 
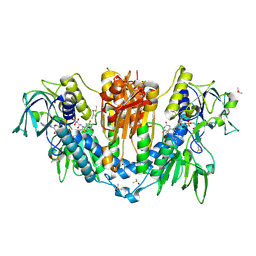 | | Trypanothione reductase | | Descriptor: | BROMIDE ION, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fiorillo, A, Ilari, A. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Innovative Approach for a Classic Target: Fragment Screening on Trypanothione Reductase Reveals New Opportunities for Drug Design.
Front Mol Biosci, 9, 2022
|
|
1R3H
 
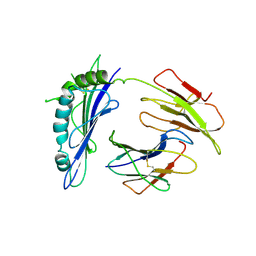 | | Crystal Structure of T10 | | Descriptor: | Beta-2-microglobulin, MHC H2-TL-T10-129 | | Authors: | Rudolph, M.G, Wilson, I.A. | | Deposit date: | 2003-10-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Combined pseudo-merohedral twinning, non-crystallographic symmetry and pseudo-translation in a monoclinic crystal form of the gammadelta T-cell ligand T10.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U9N
 
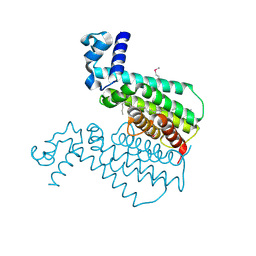 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation opens therapeutic perspectives against tuberculosis and leprosy | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
5TG6
 
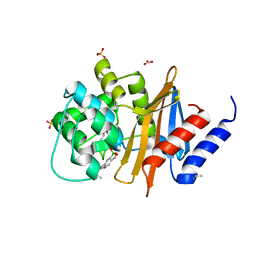 | | OXA-24/40 in Complex with Boronic Acid BA4 | | Descriptor: | BICARBONATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Powers, R.A, Werner, J.P, Mitchell, J.M. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Exploring the potential of boronic acids as inhibitors of OXA-24/40 beta-lactamase.
Protein Sci., 26, 2017
|
|
5U5Q
 
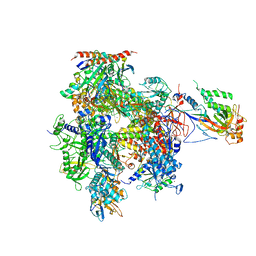 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
5TUQ
 
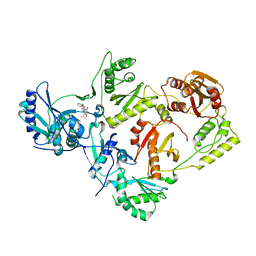 | | Crystal Structure of a 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[(benzyloxy)methyl]-6-(cyclohexylmethyl)-3-hydroxy-5-(propan-2-yl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverase transcriptase: Impacts of the 3-OH on inhibiting RNase H and polymerase.
Eur J Med Chem, 128, 2017
|
|
1R8G
 
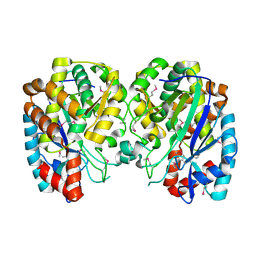 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
5TP0
 
 | | Human mesotrypsin in complex with diminazene | | Descriptor: | BERENIL, CALCIUM ION, SULFATE ION, ... | | Authors: | Kayode, O, Soares, A, Radisky, E.S. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule inhibitors of mesotrypsin from a structure-based docking screen.
PLoS ONE, 12, 2017
|
|
1U9O
 
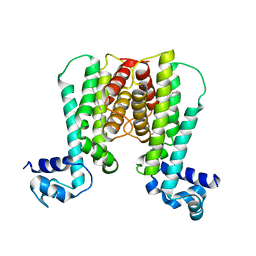 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
5TPU
 
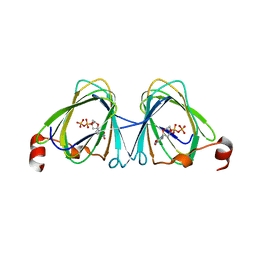 | | x-ray structure of the WlaRB TDP-quinovose 3,4-ketoisomerase from campylobacter jejuni | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Holden, H.M, Thoden, J.B, Li, J.Z, Riegert, A.S, Goneau, M.-F, Cunningham, A.M, Vinogradov, E, Schoenhofen, I.C, Gilbert, M. | | Deposit date: | 2016-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the dTDP-Fuc3N and dTDP-Qui3N biosynthetic pathways in Campylobacter jejuni 81116.
Glycobiology, 27, 2017
|
|
5UG3
 
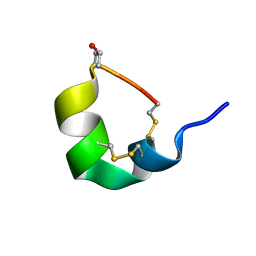 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
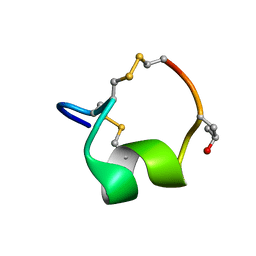 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UJ2
 
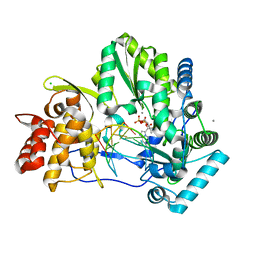 | | Crystal structure of HCV NS5B genotype 2A JFH-1 isolate with S15G E86Q E87Q C223H V321I mutations and Delta8 neta hairpoin loop deletion in complex with GS-639476 (diphsohate version of GS-9813), Mn2+ and symmetrical primer template 5'-AUAAAUUU | | Descriptor: | (1S)-1-(4-aminoimidazo[2,1-f][1,2,4]triazin-7-yl)-1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-2-methyl-D-ribitol, CHLORIDE ION, Genome polyprotein, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C, Murakami, E, Rey, A, McGrath, M.E. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1S5V
 
 | |
5SYR
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R350A | | Descriptor: | Probable ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Burgess, J.L, Burgess, R.A, Dickenson, N.E. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Spa47 Provides Mechanistic Insight into Type III Secretion System ATPase Activation and Shigella Virulence Regulation.
J. Biol. Chem., 291, 2016
|
|
1S4D
 
 | | Crystal Structure Analysis of the S-adenosyl-L-methionine dependent uroporphyrinogen-III C-methyltransferase SUMT | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, Uroporphyrin-III C-methyltransferase | | Authors: | Vevodova, J, Graham, R.M, Raux, E, Schubert, H.L, Roper, D.I, Brindley, A.A, Scott, A.I, Roessner, C.A, Stamford, N.P.J, Stroupe, M.E, Getzoff, E.D, Warren, M.J, Wilson, K.S. | | Deposit date: | 2004-01-16 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure/Function Studies on a S-Adenosyl-l-methionine-dependent Uroporphyrinogen III C Methyltransferase (SUMT), a Key Regulatory Enzyme of Tetrapyrrole Biosynthesis
J.Mol.Biol., 344, 2004
|
|
1S5W
 
 | |
5T61
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRICLINIC FORM AT 2.55 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
5T5M
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, TRIGONAL FORM AT 2.5 A. | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, GLYCEROL, HYDROSULFURIC ACID, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5TZ1
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Candida albicans in complex with the tetrazole-based antifungal drug candidate VT1161 (VT1) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Hargrove, T, Wawrzak, Z, Lepesheva, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
