4PR6
 
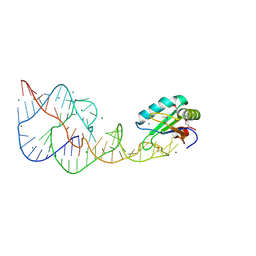 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PRF
 
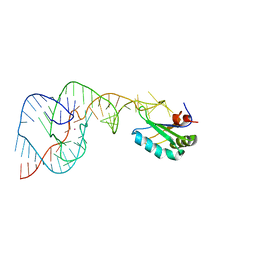 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
4W92
 
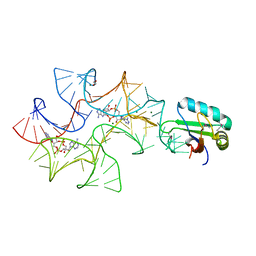 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, C-di-AMP ribsoswitch, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
4W90
 
 | | Crystal structure of Bacillus subtilis cyclic-di-AMP riboswitch ydaO | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A, ... | | Authors: | Jones, C.P, Ferre-D'Amare, A.R. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.118 Å) | | Cite: | Crystal structure of a c-di-AMP riboswitch reveals an internally pseudo-dimeric RNA.
Embo J., 33, 2014
|
|
2M9K
 
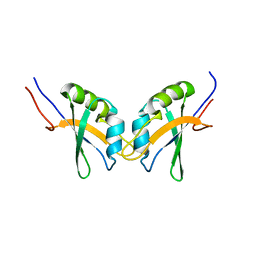 | | RBPMS2-Nter | | Descriptor: | RNA-binding protein with multiple splicing 2 | | Authors: | Yang, Y. | | Deposit date: | 2013-06-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Homodimerization of RBPMS2 through a new RRM-interaction motif is necessary to control smooth muscle plasticity.
Nucleic Acids Res., 42, 2014
|
|
4UQT
 
 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
4CH0
 
 | | RRM domain from C. elegans SUP-12 | | Descriptor: | PROTEIN SUP-12, ISOFORM B | | Authors: | Amrane, S, Mackereth, C.D. | | Deposit date: | 2013-11-27 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone-Independent Nucleic Acid Binding by Splicing Factor Sup-12 Reveals Key Aspects of Molecular Recognition
Nat.Commun., 5, 2014
|
|
2MKC
 
 | | Cooperative Structure of the Heterotrimeric pre-mRNA Retention and Splicing Complex | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-splicing factor CWC26, U2 snRNP component IST3 | | Authors: | Wysoczanski, P, Schneider, C, Xiang, S, Munari, F, Trowitzsch, S, Wahl, M.C, Luhrmann, R, Becker, S, Zweckstetter, M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CH1
 
 | |
4CIO
 
 | |
4QQB
 
 | | Structural basis for the assembly of the SXL-UNR translation regulatory complex | | Descriptor: | Protein sex-lethal, Upstream of N-ras, isoform A, ... | | Authors: | Hennig, J, Popowicz, G.M, Sattler, M. | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the assembly of the Sxl-Unr translation regulatory complex.
Nature, 515, 2014
|
|
2MGZ
 
 | | Solution structure of RBFOX family ASD-1 RRM and SUP-12 RRM in ternary complex with RNA | | Descriptor: | Protein ASD-1, isoform a, Protein SUP-12, ... | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LMZ
 
 | |
4LJM
 
 | |
2M88
 
 | |
4P6Q
 
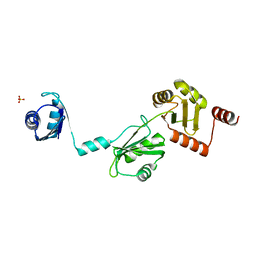 | | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA Recognition Motifs | | Descriptor: | Msx2-interacting protein, SULFATE ION | | Authors: | Arieti, F, Gabus, C, Tambalo, M, Huet, T, Round, A, Thore, S. | | Deposit date: | 2014-03-25 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the Split End protein SHARP adds a new layer of complexity to proteins containing RNA recognition motifs.
Nucleic Acids Res., 42, 2014
|
|
4N0T
 
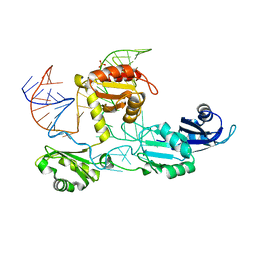 | | Core structure of the U6 small nuclear ribonucleoprotein at 1.7 Angstrom resolution | | Descriptor: | SULFATE ION, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA | | Authors: | Montemayor, E.J, Curran, E.C, Liao, H, Andrews, K.L, Treba, C.N, Butcher, S.E, Brow, D.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Core structure of the U6 small nuclear ribonucleoprotein at 1.7- angstrom resolution.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2M70
 
 | |
2RT3
 
 | | Solution structure of the second RRM domain of Nrd1 | | Descriptor: | Negative regulator of differentiation 1 | | Authors: | Kobayashi, A, Kanaba, T, Mishima, M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the second RRM domain of Nrd1, a fission yeast MAPK target RNA binding protein, and implication for its RNA recognition and regulation
Biochem.Biophys.Res.Commun., 437, 2013
|
|
2MJN
 
 | |
2MB0
 
 | |
2MKS
 
 | |
4CQ1
 
 | | Crystal structure of the neuronal isoform of PTB | | Descriptor: | CHLORIDE ION, POLYPYRIMIDINE TRACT-BINDING PROTEIN 2, ZINC ION | | Authors: | Joshi, A, Buckroyd, A.N, Curry, S. | | Deposit date: | 2014-02-10 | | Release date: | 2014-02-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Solution and Crystal Structures of a C Terminal Fragment of the Neuronal Isoform of the Polypyrimidine Tract Binding Protein (Nptb)
Peerj, 2, 2014
|
|
2MJU
 
 | |
