8V58
 
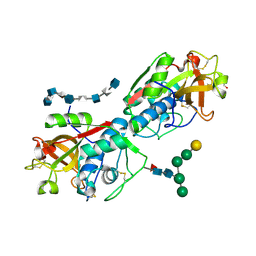 | | Complex of murine cathepsin K with bound heparan sulfate 12mer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, ... | | Authors: | Pedersen, L.C, Xu, D, Krahn, J.M. | | Deposit date: | 2023-11-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Heparan sulfate selectively inhibits the collagenase activity of cathepsin K.
Matrix Biol., 129, 2024
|
|
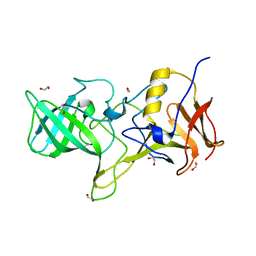
4OOS
 
 | |
6URC
 
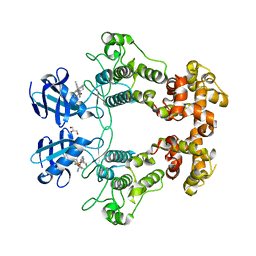 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7GKZ
 
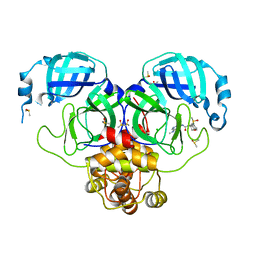 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-afb6844f-1 (Mpro-P1474) | | Descriptor: | (4R)-6-chloro-N-[4-methyl-5-(methylamino)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3EJT
 
 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog:(5R)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5R,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
3EKN
 
 | | Insulin receptor kinase complexed with an inhibitor | | Descriptor: | 2-fluoro-6-{[2-({2-methoxy-4-[4-(1-methylethyl)piperazin-1-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}benzamide, Insulin receptor | | Authors: | Chamberlain, S, Atkins, C, Deanda, F, Dumble, M, Gerding, R, Groy, A, Korenchuk, S, Kumar, R, Lei, H, Mook, R, Moorthy, G, Redman, A, Rowland, J, Shewchuk, L. | | Deposit date: | 2008-09-19 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 4,6-bis-anilino-1H-pyrrolo[2,3-d]pyrimidine IGF-1R tyrosine kinase inhibitors towards JNK selectivity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6S05
 
 | |
5WMA
 
 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
5V96
 
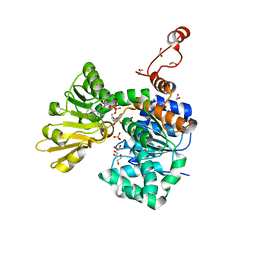 | |
7GKH
 
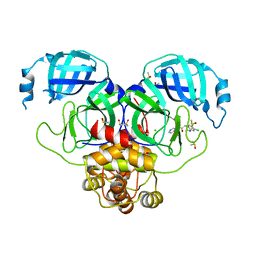 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-UNI-8d415491-1 (Mpro-P0884) | | Descriptor: | (4R)-6,7-dichloro-N-(4-cyclopropylpyridin-3-yl)-1,2,3,4-tetrahydroquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3GWT
 
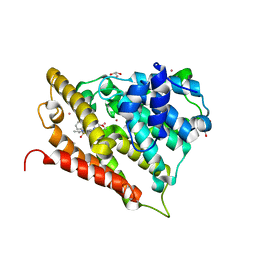 | |
9LXL
 
 | | Crystal structure of GH29 family alpha-L-fucosidase from Fusarium proliferatum LE1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Korban, S.A, Borshchevskiy, V.I, Pospelov, V.A, Bobrov, K.S, Ivanova, D.N, Kulminskaya, A.A. | | Deposit date: | 2025-02-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structure and biochemical characterization of GH29 family alpha-l-fucosidase from Fusarium proliferatum LE1.
Biochem.Biophys.Res.Commun., 779, 2025
|
|
6C1W
 
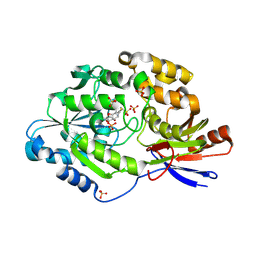 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemase, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-01-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Biochemistry, 57, 2018
|
|
5WF5
 
 | | Agonist bound human A2a adenosine receptor with D52N mutation at 2.60 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Human A2a adenosine receptor T4L chimera | | Authors: | White, K.L, Eddy, M.T, Gao, Z, Han, G.W, Hanson, M.A, Lian, T, Deary, A, Patel, N, Jacobson, K.A, Katritch, V, Stevens, R.C. | | Deposit date: | 2017-07-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling.
Structure, 26, 2018
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
6S6Y
 
 | | X-ray crystal structure of the formyltransferase/hydrolase complex (FhcABCD) from Methylorubrum extorquens in complex with methylofuran | | Descriptor: | (2~{S})-3-[4-[[5-(aminomethyl)furan-3-yl]methoxy]phenyl]-2-(methylamino)propanoic acid, 1,2-ETHANEDIOL, AMINO GROUP, ... | | Authors: | Wagner, T, Hemmann, J.L, Shima, S, Vorholt, J. | | Deposit date: | 2019-07-04 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7GLR
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MIC-UNK-08cd9c58-1 (Mpro-P2010) | | Descriptor: | 2-(3-chlorophenyl)-N-(1,7-naphthyridin-5-yl)acetamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
3EJS
 
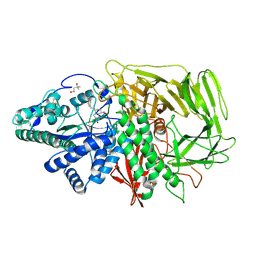 | | Golgi alpha-Mannosidase II in complex with 5-substituted swainsonine analog: (5S)-5-[2'-(4-tert-butylphenyl)ethyl]-swainsonine | | Descriptor: | (1S,2R,5S,8R,8aR)-5-[2-(4-tert-butylphenyl)ethyl]octahydroindolizine-1,2,8-triol, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Shea, K, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
6MIA
 
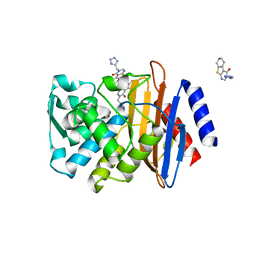 | | Crystal structure of CTX-M-14 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-09-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
8Y39
 
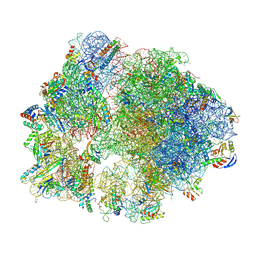 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
6MIS
 
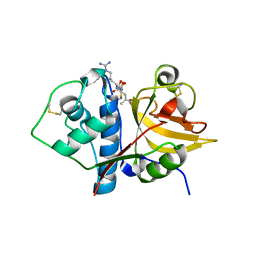 | | Native ananain in complex with E-64 | | Descriptor: | Ananain, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Yongqing, T, Wilmann, P.G, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Determination of the crystal structure and substrate specificity of ananain.
Biochimie, 166, 2019
|
|
3E8S
 
 | |
5CT2
 
 | | The structure of the NK1 fragment of HGF/SF complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
3E88
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-1,2,5-oxadiazol-3-yl)-6-{[(2R)-2-amino-3-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
