3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3HID
 
 | | Crystal structure of adenylosuccinate synthetase from Yersinia pestis CO92 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Adenylosuccinate synthetase | | Authors: | Zhang, R, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the adenylosuccinate synthetase from Yersinia pestis CO92
To be Published
|
|
6E8T
 
 | |
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
6Q17
 
 | | Crystal structure of Human galectin-3 CRD in complex with Methyl 3-O-[1-(b-D-galactopyranosyl)-1,2,3-triazol-4-yl]-methyl-b-D-galactopyranoside | | Descriptor: | CHLORIDE ION, Galectin-3, methyl 3-O-[(1-beta-D-galactopyranosyl-1H-1,2,3-triazol-4-yl)methyl]-beta-D-galactopyranoside | | Authors: | Kishor, C, Blanchard, H. | | Deposit date: | 2019-08-02 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Linear triazole-linked pseudo oligogalactosides as scaffolds for galectin inhibitor development.
Chem.Biol.Drug Des., 96, 2020
|
|
7FCC
 
 | | IL-1RAcPb TIR domain | | Descriptor: | Isoform 4 of Interleukin-1 receptor accessory protein | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
4PG3
 
 | | Crystal structure of KRS complexed with inhibitor | | Descriptor: | LYSINE, Lysine--tRNA ligase, cladosporin | | Authors: | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
8W7H
 
 | | Purine Nucleoside Phosphorylase in complex with MMV000848 | | Descriptor: | (2R)-1-(9H-carbazol-9-yl)-3-(cyclopentylamino)propan-2-ol, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Chung, Z, Lin, J.Q, Lescar, J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and structural validation of purine nucleoside phosphorylase from Plasmodium falciparum as a target of MMV000848.
J.Biol.Chem., 300, 2023
|
|
2VUJ
 
 | | Environmentally isolated GH11 xylanase | | Descriptor: | GH11 XYLANASE, GLYCEROL | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, DeSantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
7B9Y
 
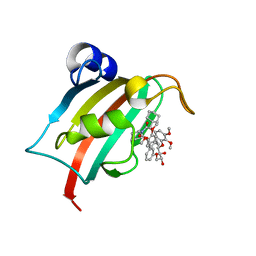 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
5T9R
 
 | | Structure of rabbit RyR1 (Ca2+-only dataset, class 3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6GRG
 
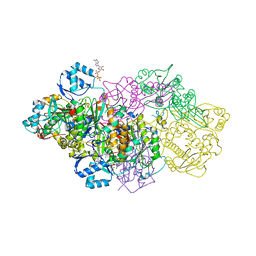 | | E. coli Microcin synthetase McbBCD complex with pro-MccB17, ADP and phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ghilarov, D, Stevenson, C.E.M, Travin, D.Y, Piskunova, J, Serebryakova, M, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of Microcin B17 Synthetase: An Octameric Protein Complex Converting a Ribosomally Synthesized Peptide into a DNA Gyrase Poison.
Mol. Cell, 73, 2019
|
|
6ADF
 
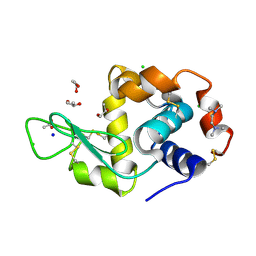 | | Structure of HEWL co-crystallised with TEMED | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Seyedarabi, A, Seraj, Z. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The aroma of TEMED as an activation and stabilizing signal for the antibacterial enzyme HEWL.
Plos One, 15, 2020
|
|
5TB0
 
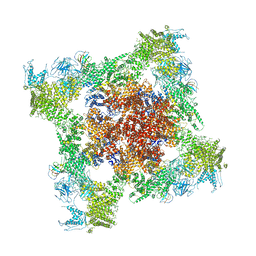 | | Structure of rabbit RyR1 (EGTA-only dataset, all particles) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
3HFW
 
 | | Crystal Structure of human ADP-ribosylhydrolase 1 (hARH1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S, Mueller-Dieckmann, J, Koch-Nolte, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of human ADP-ribosylhydrolase 1
To be Published
|
|
8HUI
 
 | | Crystal structure of DFA I-forming Inulin Lyase from Streptomyces peucetius subsp. caesius ATCC 27952 in complex with GF4, DFA I, and fructose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cheng, M, Rao, Y.J, Mu, W.M. | | Deposit date: | 2022-12-24 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insights into the Catalytic Cycle of Inulin Fructotransferase: From Substrate Anchoring to Product Releasing.
J.Agric.Food Chem., 72, 2024
|
|
7X5P
 
 | | Truncated VhChiP (1-19aa) in complex with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Robinson, C.R, Aunkham, A, Suginta, W. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of truncated VhChiP
To Be Published
|
|
7B9Z
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 35-(E) | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,20,24(28),25-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5, isothiocyanate | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4C6L
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to the inhibitor fluoroorotate at pH 6.0 | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
7IBD
 
 | |
7IBI
 
 | |
4C9I
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 1E protein (Form B) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, GLYCEROL, MAJOR STRAWBERRY ALLERGEN FRA A 1-E | | Authors: | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
3ERD
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
9HCU
 
 | | Crystal structure of human TRF1 TRFH domain in complex with compound 51 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-(4-fluorophenyl)-2-(piperazin-1-yl)acetamide, ... | | Authors: | Casale, G, Le Bihan, Y.-V, van Montfort, R.L.M, Guettler, S. | | Deposit date: | 2024-11-11 | | Release date: | 2025-11-19 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of first-in-class inhibitors of the TRF1-TIN2 protein-protein interaction by fragment screening
To Be Published
|
|
7IBL
 
 | |
