6R5D
 
 | |
6R4U
 
 | |
6XEO
 
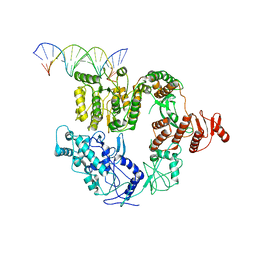 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
6PFJ
 
 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-21 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
2PX5
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Orthorhombic crystal form) | | Descriptor: | Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
5V37
 
 | | Crystal structure of SMYD3 with SAM and EPZ028862 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Small molecule inhibitors and CRISPR/Cas9 mutagenesis demonstrate that SMYD2 and SMYD3 activity are dispensable for autonomous cancer cell proliferation.
Plos One, 13, 2018
|
|
4L5G
 
 | | Crystal structure of Thermus thermophilus CarD | | Descriptor: | CarD | | Authors: | Srivastava, D.B, Westblade, L.F, Campbell, E.A, Darst, S.A. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3902 Å) | | Cite: | Structure and function of CarD, an essential mycobacterial transcription factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6R4S
 
 | |
6R4T
 
 | |
6RB4
 
 | |
6R5E
 
 | |
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
2PX4
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Monoclinic form 2) | | Descriptor: | CHLORIDE ION, GLYCEROL, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PXA
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and GTPG | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PX2
 
 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH (Monoclinic form 1) | | Descriptor: | CHLORIDE ION, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
6IC9
 
 | |
2C5Z
 
 | | Structure and CTD binding of the Set2 SRI domain | | Descriptor: | SET DOMAIN PROTEIN 2 | | Authors: | Vojnic, E, Simon, B, Strahl, B.D, Sattler, M, Cramer, P. | | Deposit date: | 2005-11-03 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription.
J. Biol. Chem., 281, 2006
|
|
2PX8
 
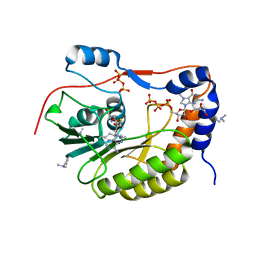 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and 7M-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
2PXC
 
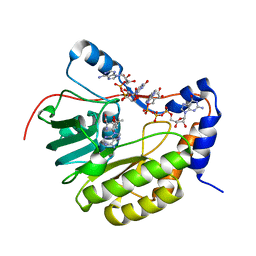 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAM and GTPA | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYLMETHIONINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
4N4I
 
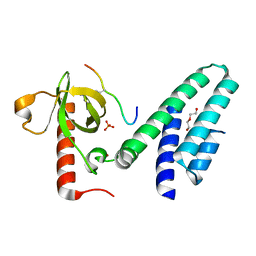 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
5DZM
 
 | |
5FVC
 
 | | Structure of RNA-bound decameric HMPV nucleoprotein | | Descriptor: | HMPV NUCLEOPROTEIN, RNA | | Authors: | Renner, M, Bertinelli, M, Leyrat, C, Paesen, G.C, Saraiva de Oliveira, L.F, Huiskonen, J.T, Grimes, J.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein.
Elife, 5, 2016
|
|
6FSB
 
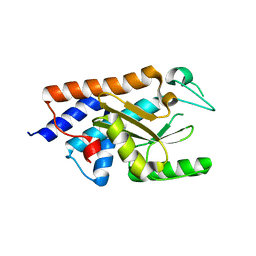 | | Influenza B/Memphis/13/03 endonuclease with I38T mutation | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Cusack, S, Speranzini, V. | | Deposit date: | 2018-02-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil.
Sci Rep, 8, 2018
|
|
4NQW
 
 | |
4TRQ
 
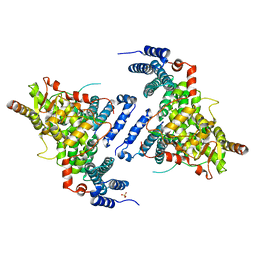 | | Crystal structure of Sac3/Thp1/Sem1 | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Hellerschmied, D, Schneider, S, Kohler, A, Clausen, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.
Cell, 162, 2015
|
|
