5ZSL
 
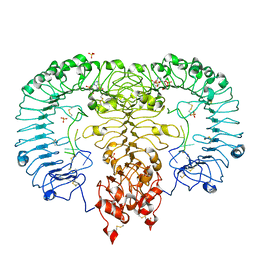 | | Crystal structure of monkey TLR7 in complex with GGUUGG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2S,3aR,4R,6R,6aR)-2-hydroxy-6-(hydroxymethyl)-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]-3,9-dihydro-6H-purin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSE
 
 | | Crystal structure of monkey TLR7 in complex with IMDQ and GGUCCC | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
6GHT
 
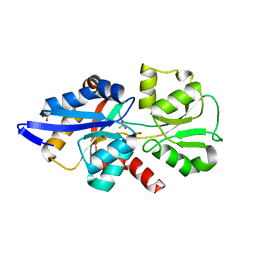 | | HtxB D206A protein variant from Pseudomonas stutzeri in complex with hypophosphite to 1.12 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
5ZSD
 
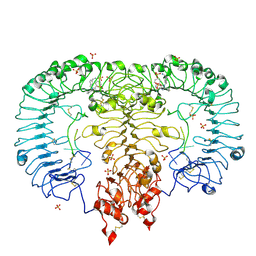 | | Crystal structure of monkey TLR7 in complex with IMDQ and GGUUGG | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSM
 
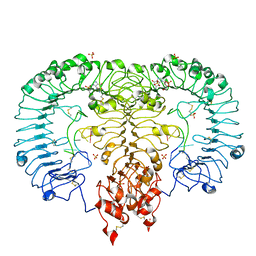 | | Crystal structure of monkey TLR7 in complex with GGUCCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2S,3aR,4R,6R,6aR)-2-hydroxy-6-(hydroxymethyl)-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]-3,9-dihydro-6H-purin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSB
 
 | | Crystal structure of monkey TLR7 in complex with IMDQ and AAUUAA | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSF
 
 | | Crystal structure of monkey TLR7 in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSA
 
 | | Crystal structure of monkey TLR7 in complex with IMDQ and UUUUUU | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSJ
 
 | | Crystal structure of monkey TLR7 in complex with GS9620 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-azanyl-2-butoxy-8-[[3-(pyrrolidin-1-ylmethyl)phenyl]methyl]-5,7-dihydropteridin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
5ZSG
 
 | | Crystal structure of monkey TLR7 in complex with gardiquimod | | Descriptor: | 1-{4-amino-2-[(ethylamino)methyl]-1H-imidazo[4,5-c]quinolin-1-yl}-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
3CDS
 
 | |
5ZSC
 
 | | Crystal structure of monkey TLR7 in complex with IMDQ and CCUUCC | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
3PCD
 
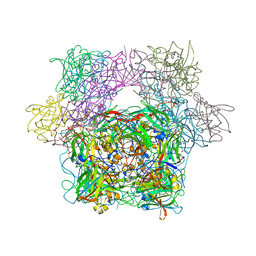 | | PROTOCATECHUATE 3,4-DIOXYGENASE Y447H MUTANT | | Descriptor: | BETA-MERCAPTOETHANOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
Biochemistry, 37, 1998
|
|
3PCI
 
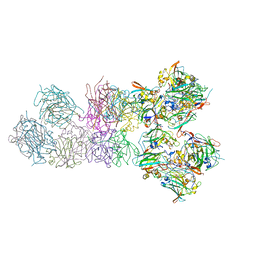 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-IODO-4-HYDROXYBENZOATE | | Descriptor: | 3-IODO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3PCH
 
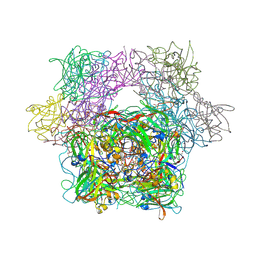 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-CHLORO-4-HYDROXYBENZOATE | | Descriptor: | 3-CHLORO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3CA7
 
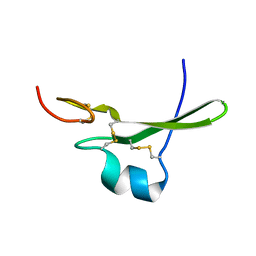 | |
8E7X
 
 | | RsTSPO A138F with one Heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7Y
 
 | | RsTSPO A138F with two heme bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
6ECQ
 
 | | The human methylenetetrahydrofolate dehydrogenase/cyclohydrolase (FolD) complexed with NADP and inhibitor LY345899 | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE CYCLOHYDROLASE, N-{4-[(6aR)-3-amino-1,9-dioxo-1,2,5,6,6a,7-hexahydroimidazo[1,5-f]pteridin-8(9H)-yl]benzene-1-carbonyl}-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bueno, R.V, Dawson, A, Hunter, W.N. | | Deposit date: | 2018-08-08 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An assessment of three human methylenetetrahydrofolate dehydrogenase/cyclohydrolase-ligand complexes following further refinement.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
8E7Z
 
 | | RsTSPO mutant -A138F | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Tryptophan-rich sensory protein | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
8E7W
 
 | | RsTSPO A139T with Heme | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Liu, J, Hiser, C, Li, F, Garavito, R, Ferguson-Miller, S. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New TSPO Crystal Structures of Mutant and Heme-Bound Forms with Altered Flexibility, Ligand Binding, and Porphyrin Degradation Activity.
Biochemistry, 62, 2023
|
|
4PFF
 
 | | Crystal structure of Plasmodium vivax SHMT with PLP Schiff base | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, putative | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2014-04-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Plasmodium vivax serine hydroxymethyltransferase: implications for ligand-binding specificity and functional control.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PCF
 
 | | STRUCTURE OF PROTOCATECHUATE 3,4-DIOXYGENASE COMPLEXED WITH 3-FLURO-4-HYDROXYBENZOATE | | Descriptor: | 3-FLUORO-4-HYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, FE (III) ION, ... | | Authors: | Orville, A.M, Elango, N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-06-27 | | Release date: | 1998-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of competitive inhibitor complexes of protocatechuate 3,4-dioxygenase: multiple exogenous ligand binding orientations within the active site.
Biochemistry, 36, 1997
|
|
3C9A
 
 | | High Resolution Crystal Structure of Argos bound to the EGF domain of Spitz | | Descriptor: | BROMIDE ION, Protein giant-lens, Protein spitz | | Authors: | Klein, D.E, Stayrook, S.E, Shi, F, Narayan, K, Lemmon, M.A. | | Deposit date: | 2008-02-15 | | Release date: | 2008-05-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for EGFR ligand sequestration by Argos.
Nature, 453, 2008
|
|
