2OKD
 
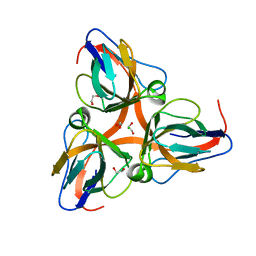 | |
2OKB
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OL1
 
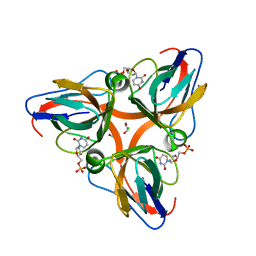 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6TE4
 
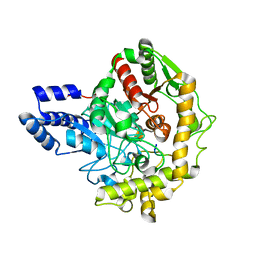 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | Descriptor: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | Authors: | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
5FWK
 
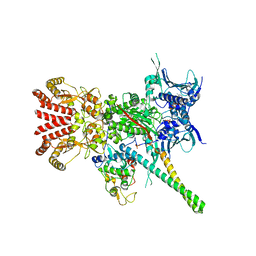 | | Atomic cryoEM structure of Hsp90-Cdc37-Cdk4 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CYCLIN-DEPENDENT KINASE 4, HEAT SHOCK PROTEIN HSP 90 BETA, ... | | Authors: | Verba, K.A, Wang, R.Y.R, Arakawa, A, Liu, Y, Yokoyama, S, Agard, D.A. | | Deposit date: | 2016-02-17 | | Release date: | 2016-07-06 | | Last modified: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic Structure of Hsp90-Cdc37-Cdk4 Reveals that Hsp90 Traps and Stabilizes an Unfolded Kinase.
Science, 352, 2016
|
|
6TEB
 
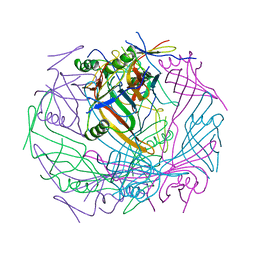 | | Tail-baseplate interface of native GTA particle computed with C6 symmetry | | Descriptor: | Distal tail protein Rcc01695, Tail tube protein Rcc01691 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6FIB
 
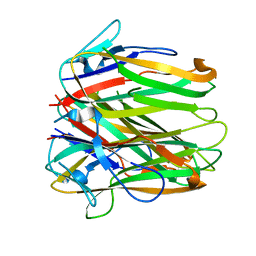 | | Structure of human 4-1BB ligand | | Descriptor: | Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor ligand superfamily member 9,4-1BBL -CH/CL fusion, Tumor necrosis factor ligand superfamily member 9,Uncharacterized protein | | Authors: | Joseph, C, Claus, C, Ferrara, C, von Hirschheydt, T, Prince, C, Funk, D, Klein, C, Benz, J. | | Deposit date: | 2018-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tumor-targeted 4-1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy.
Sci Transl Med, 11, 2019
|
|
6T63
 
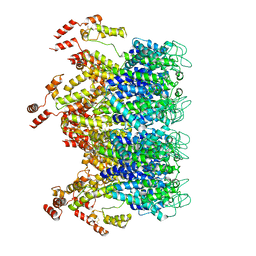 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
7LDK
 
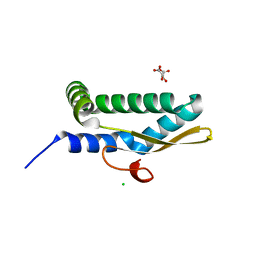 | | Structure of human respiratory syncytial virus nonstructural protein 2 (NS2) | | Descriptor: | CHLORIDE ION, D(-)-TARTARIC ACID, Non-structural protein 2 | | Authors: | Chatterjee, S, Borek, D, Otwinowski, Z, Amarasinghe, G.K, Leung, D.W. | | Deposit date: | 2021-01-13 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for IFN antagonism by human respiratory syncytial virus nonstructural protein 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPB
 
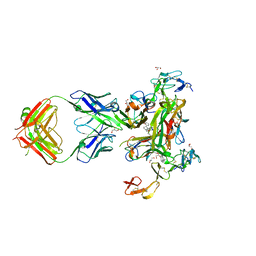 | | Human TNF-alpha TNFR1 complex bound to conformationally selective antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(1-{[2-(difluoromethoxy)phenyl]methyl}-2-{[3-(2-oxopyrrolidin-1-yl)phenoxy]methyl}-1H-benzimidazol-6-yl)pyridin-2(1H)-one, Fab1974 - Heavy Chain, ... | | Authors: | Fox III, D, Conrady, D.G, Lowe, M, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF
Nat Commun, 12, 2021
|
|
4D46
 
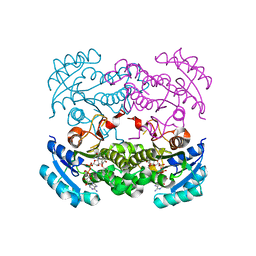 | | Crystal structure of E. coli FabI in complex with NAD and 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile | | Descriptor: | 5-bromo-2-(4-chloro-2-hydroxyphenoxy)benzonitrile, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tareilus, M, Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
4D42
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 4-fluoro- 5-hexyl-2-phenoxyphenol | | Descriptor: | 4-fluoro-5-hexyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An ordered water channel in Staphylococcus aureus FabI: unraveling the mechanism of substrate recognition and reduction.
Biochemistry, 54, 2015
|
|
4B6O
 
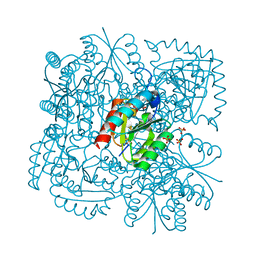 | | Structure of Mycobacterium tuberculosis Type II Dehydroquinase inhibited by (2S)-2-(4-methoxy)benzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(4-methoxyphenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
8ODZ
 
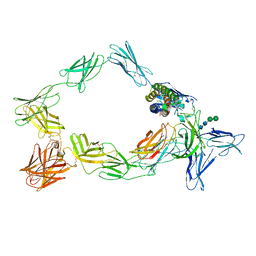 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4GJT
 
 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
8OE4
 
 | | Cryo-EM structure of a pre-dimerized human IL-23 complete extracellular signaling complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Bloch, Y, Felix, J, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OE0
 
 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 2). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-03-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4Q3K
 
 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
2JLL
 
 | | Crystal structure of NCAM2 IgIV-FN3II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Kulahin, N, Rasmussen, K, Kristensen, O, Kastrup, J, Berezin, V, Bock, E, Walmod, P, Gajhede, M. | | Deposit date: | 2008-09-10 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Model and Trans-Interaction of the Entire Ectodomain of the Olfactory Cell Adhesion Molecule.
Structure, 19, 2011
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
3MS6
 
 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
3IFD
 
 | |
2WB5
 
 | | GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation | | Descriptor: | (5R,6R,7R,8S)-6,7-dihydroxy-5-(hydroxymethyl)-2-(2-phenylethyl)-8-(propanoylamino)-5,6,7,8-tetrahydro-1H-imidazo[1,2-a]pyridin-4-ium, CHLORIDE ION, O-GLCNACASE NAGJ, ... | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Glcnacstatins are Nanomolar Inhibitors of Human O-Glcnacase Inducing Cellular Hyper-O-Glcnacylation
Biochem.J., 420, 2009
|
|
3S9L
 
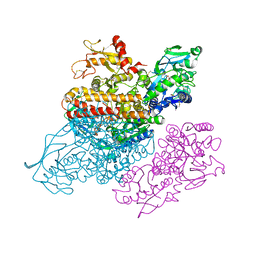 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6MGP
 
 | | Structure of human 4-1BB / 4-1BBL complex | | Descriptor: | ACETATE ION, GLYCEROL, Tumor necrosis factor ligand superfamily member 9, ... | | Authors: | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab
Nat Commun, 9, 2018
|
|
