4E5L
 
 | | Crystal structure of avian influenza virus PAn bound to compound 6 | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5H
 
 | | Crystal structure of avian influenza virus PAn bound to compound 3 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5G
 
 | | Crystal structure of avian influenza virus PAn bound to compound 2 | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5E
 
 | | Crystal structure of avian influenza virus PAn Apo | | Descriptor: | MANGANESE (II) ION, Polymerase protein PA, SULFATE ION | | Authors: | DuBois, R.M, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5F
 
 | | Crystal structure of avian influenza virus PAn bound to compound 1 | | Descriptor: | 2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
4E5I
 
 | | Crystal structure of avian influenza virus PAn bound to compound 4 | | Descriptor: | 5-hydroxy-2-(1-methyl-1H-imidazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxylic acid, MANGANESE (II) ION, Polymerase protein PA, ... | | Authors: | DuBois, R.M, Slavish, P.J, Webb, T.R, White, S.W. | | Deposit date: | 2012-03-14 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.944 Å) | | Cite: | Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease.
Plos Pathog., 8, 2012
|
|
7KTR
 
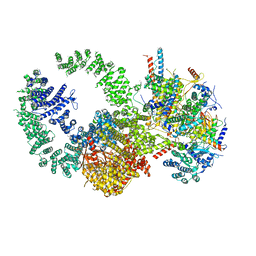 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7K5I
 
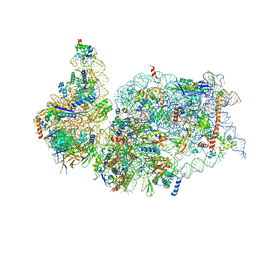 | | SARS-COV-2 nsp1 in complex with human 40S ribosome | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Wang, L, Shi, M, Wu, H. | | Deposit date: | 2020-09-16 | | Release date: | 2020-10-14 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism.
Biorxiv, 2020
|
|
3VU3
 
 | |
6FS7
 
 | |
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6WQD
 
 | | The 1.95 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Kim, Y, Wilamowski, M, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
5MG2
 
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
2FQM
 
 | |
6FS6
 
 | |
2ETN
 
 | | Crystal structure of Thermus aquaticus Gfh1 | | Descriptor: | anti-cleavage anti-GreA transcription factor Gfh1 | | Authors: | Lamour, V, Hogan, B.P, Erie, D.A, Darst, S.A. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage.
J.Mol.Biol., 356, 2006
|
|
6TB4
 
 | | Structure of SAGA bound to TBP | | Descriptor: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
1VPT
 
 | | AS11 VARIANT OF VACCINIA VIRUS PROTEIN VP39 IN COMPLEX WITH S-ADENOSYL-L-METHIONINE | | Descriptor: | S-ADENOSYLMETHIONINE, VP39 | | Authors: | Hodel, A.E, Gershon, P.D, Shi, X, Quiocho, F.A. | | Deposit date: | 1996-03-20 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends.
Cell(Cambridge,Mass.), 85, 1996
|
|
5G5P
 
 | | Structure of the Saccharomyces cerevisiae TREX-2 complex | | Descriptor: | 26S PROTEASOME COMPLEX SUBUNIT SEM1, NUCLEAR MRNA EXPORT PROTEIN SAC3, NUCLEAR MRNA EXPORT PROTEIN THP1 | | Authors: | Aibara, S, Bai, X.C, Stewart, M. | | Deposit date: | 2016-05-26 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | The Sac3 Tpr-Like Region in the Saccharomyces Cerevisiae Trex-2 Complex is More Extensive But Independent of the Cid Region
J.Struct.Biol., 195, 2016
|
|
2HMI
 
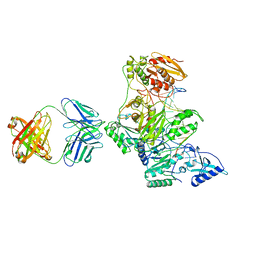 | | HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3'), FAB FRAGMENT OF MONOCLONAL ANTIBODY 28, ... | | Authors: | Ding, J, Arnold, E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution.
J.Mol.Biol., 284, 1998
|
|
8BGU
 
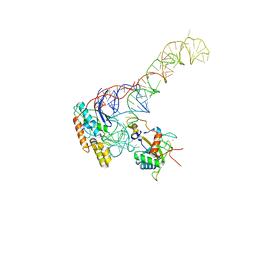 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6P1B
 
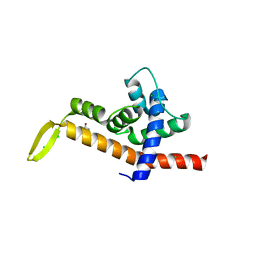 | | Transcription antitermination factor Q21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Q protein | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
6P1A
 
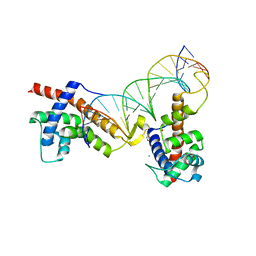 | |
