4CBP
 
 | | Crystal structure of neural ectodermal development factor IMP-L2. | | Descriptor: | GLYCEROL, NEURAL/ECTODERMAL DEVELOPMENT FACTOR IMP-L2 | | Authors: | Kulahin, N, Kristensen, O, Brzozowski, M, Schluckebier, G, Meyts, P.D. | | Deposit date: | 2013-10-15 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Analysis of Imp-L2 Function
To be Published
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3ZDG
 
 | | Crystal Structure of Ls-AChBP complexed with carbamoylcholine analogue 3-(dimethylamino)butyl dimethylcarbamate (DMABC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(dimethylamino)butyl dimethylcarbamate, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
3ZSJ
 
 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZLV
 
 | | Crystal structure of mouse acetylcholinesterase in complex with tabun and HI-6 | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
3ZSK
 
 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZLT
 
 | | Crystal structure of acetylcholinesterase in complex with RVX | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
4EGU
 
 | | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ZINC ION, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-01 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | 0.95A Resolution Structure of a Histidine Triad Protein from Clostridium difficile
To be Published
|
|
4COJ
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima in complex with dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4BTL
 
 | | Aromatic interactions in acetylcholinesterase-inhibitor complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Qian, W, Engdahl, C, Berg, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2013-06-18 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4COI
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with glycerol in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, GLYCEROL, ZINC ION | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
3KPU
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-quinolinol | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-4-ol | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQM
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-Bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3L4E
 
 | |
3KQO
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chloropurine | | Descriptor: | 6-chloro-9H-purine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3L8X
 
 | | P38 alpha kinase complexed with a pyrazolo-pyrimidine based inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N,4-dimethyl-3-[(1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Sack, J.S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pyrazolo-Pyrimidines: A Novel Heterocyclic Scaffold for Potent and Selective P38 Alpha Inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3LU2
 
 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
3UVR
 
 | | Human p38 MAP Kinase in Complex with KM064 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{3-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
3V03
 
 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3UVQ
 
 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
3M8A
 
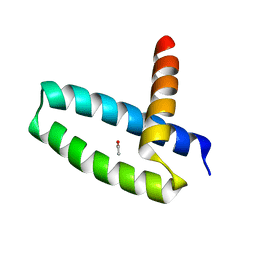 | | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009 | | Descriptor: | ACETATE ION, MALONATE ION, Nonstructural protein 1, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009
To be Published
|
|
3KPV
 
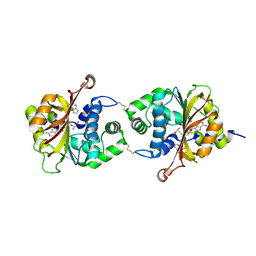 | | Crystal Structure of hPNMT in Complex AdoHcy and Adenine | | Descriptor: | ADENINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3KQQ
 
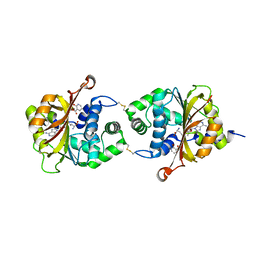 | | Crystal Structure of hPNMT in Complex AdoHcy and 2-Hydroxynicotinic acid | | Descriptor: | 2-oxo-1,2-dihydropyridine-3-carboxylic acid, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
3UEE
 
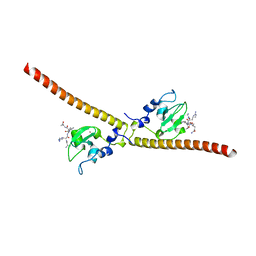 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
