1IXC
 
 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-06-18 | | Release date: | 2003-06-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IZ1
 
 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | Descriptor: | LysR-type regulatory protein | | Authors: | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | Deposit date: | 2002-09-18 | | Release date: | 2003-05-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
5AE4
 
 | | Structures of inactive and activated DntR provide conclusive evidence for the mechanism of action of LysR transcription factors | | Descriptor: | LYSR-TYPE REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Lerche, M, Dian, C, Round, A, Lonneborg, R, Brzezinski, P, Leonard, G.A. | | Deposit date: | 2015-08-25 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Solution Configurations of Inactive and Activated Dntr Have Implications for the Sliding Dimer Mechanism of Lysr Transcription Factors.
Sci.Rep., 6, 2016
|
|
9HH1
 
 | | LysR Type Transcriptional Regulator LsrB from Agrobacterium tumefaciens | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Elders, H, Schmidt, J.J, Fiedler, R, Hofmann, E, Narberhaus, F. | | Deposit date: | 2024-11-20 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Two redox-responsive LysR-type transcription factors control the oxidative stress response of Agrobacterium tumefaciens.
Nucleic Acids Res., 53, 2025
|
|
3T1B
 
 | | Crystal structure of the full-length AphB N100E variant | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
3SZP
 
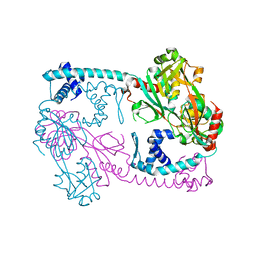 | | Full-length structure of the Vibrio cholerae virulence activator, AphB, a member of the LTTR protein family | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
5AE5
 
 | | Structures of inactive and activated DntR provide conclusive evidence for the mechanism of action of LysR transcription factors | | Descriptor: | GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Lerche, M, Dian, C, Round, A, Lonneborg, R, Brzezinski, P, Leonard, G.A. | | Deposit date: | 2015-08-25 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | The Solution Configurations of Inactive and Activated Dntr Have Implications for the Sliding Dimer Mechanism of Lysr Transcription Factors.
Sci.Rep., 6, 2016
|
|
8Y6U
 
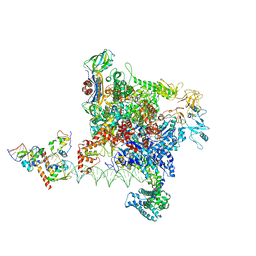 | |
1UTB
 
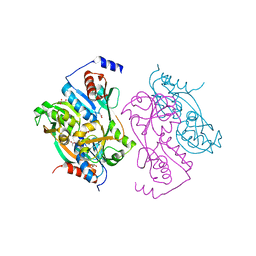 | | DntR from Burkholderia sp. strain DNT | | Descriptor: | ACETATE ION, GLYCEROL, LYSR-TYPE REGULATORY PROTEIN | | Authors: | Smirnova, I.A, Dian, C, Leonard, G.A, McSweeney, S, Birse, D, Brzezinski, P. | | Deposit date: | 2003-12-05 | | Release date: | 2004-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of a Bacterial Biosensor for Nitrotoluenes: The Crystal Structure of the Transcriptional Regulator Dntr
J.Mol.Biol., 340, 2004
|
|
9FDD
 
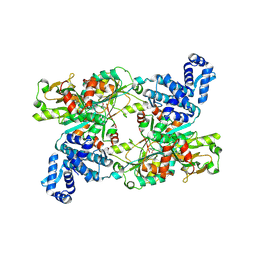 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-05-16 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
9F14
 
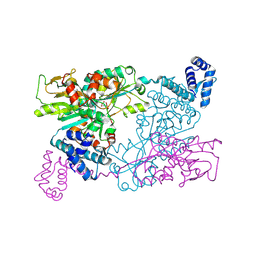 | | The crystal structure of full length tetramer CysB from Klebsiella aerogenes in complex with N-acetylserine | | Descriptor: | HTH-type transcriptional regulator CysB, N-ACETYL-SERINE | | Authors: | Verschueren, K.H.G, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2024-04-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the LysR-type Transcriptional Regulator, CysB, Bound to the Inducer, N-acetylserine.
Eur.Biophys.J., 53, 2024
|
|
1UTH
 
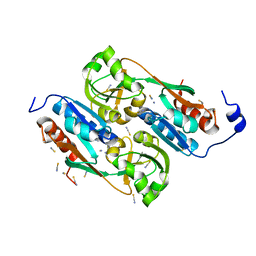 | | DntR from Burkholderia sp. strain DNT in complex with Thiocyanate | | Descriptor: | LYSR-TYPE REGULATORY PROTEIN, THIOCYANATE ION | | Authors: | Smirnova, I.A, Dian, C, Leonard, G.A, McSweeney, S, Birse, D, Brzezinski, P. | | Deposit date: | 2003-12-09 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of a Bacterial Biosensor for Nitrotoluenes: The Crystal Structure of the Transcriptional Regulator Dntr
J.Mol.Biol., 340, 2004
|
|
3MZ1
 
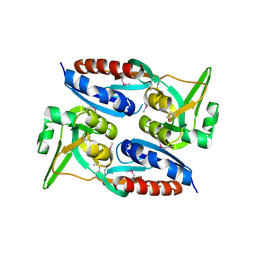 | | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021 | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021
To be Published
|
|
6G1B
 
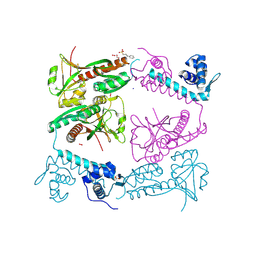 | | Corynebacterium glutamicum OxyR, oxidized form | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, HYDROGEN PEROXIDE, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G4R
 
 | | Corynebacterium glutamicum OxyR C206S mutant, H2O2-bound | | Descriptor: | 1,2-ETHANEDIOL, HYDROGEN PEROXIDE, Hydrogen peroxide-inducible genes activator, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-28 | | Release date: | 2018-12-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G1D
 
 | | Corynebacterium glutamicum OxyR C206 mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7QA0
 
 | |
7QA3
 
 | |
7QAV
 
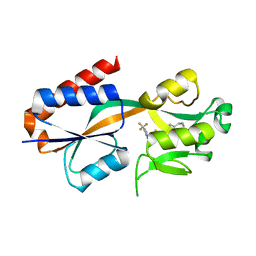 | |
7W06
 
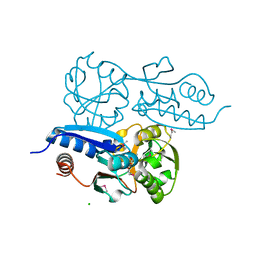 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W07
 
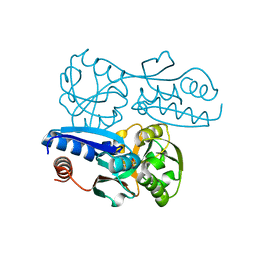 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W08
 
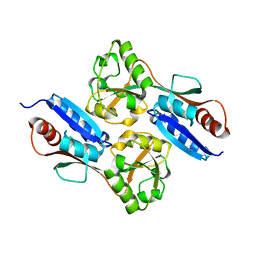 | |
3FXQ
 
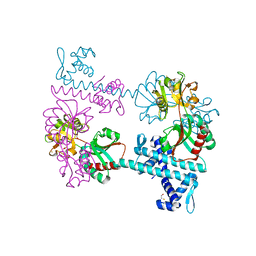 | | Crystal structure of the LysR-type transcriptional regulator TsaR | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3FXR
 
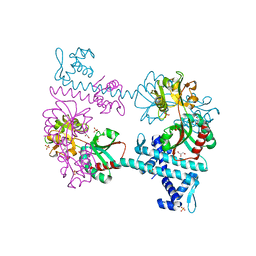 | | Crystal structure of TsaR in complex with sulfate | | Descriptor: | ASCORBIC ACID, GLYCEROL, LysR type regulator of tsaMBCD, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
