6N7F
 
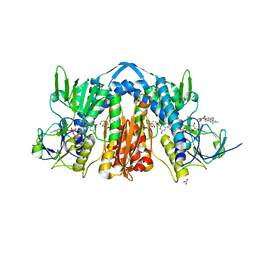 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-27 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
8A4D
 
 | |
6QXY
 
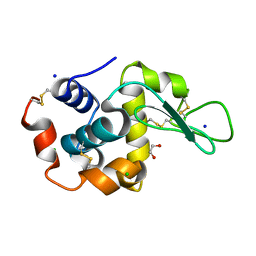 | | Pink beam serial crystallography: Lysozyme, 5 us exposure, 8813 patterns merged (1 chip) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
6QY4
 
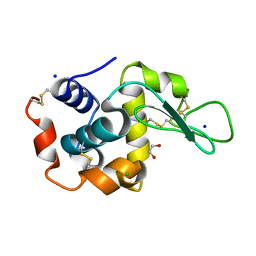 | | Pink beam serial crystallography: Lysozyme, 1 us exposure, 14793 patterns merged | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
5KRY
 
 | |
5KQO
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, PHOSPHATE ION | | Authors: | Ussin, N, Offermann, L.R, Perdue, M, Chruszcz, M. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
5KS1
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, ... | | Authors: | Ussin, N, Abdulsalam, R.W, Offermann, L.R, Chruszcz, M. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
5KRR
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus in complex with Mn(2+) | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, CHLORIDE ION, ... | | Authors: | Ussin, N, Abdulsalam, R.W, Offermann, L.R, Perdue, M, Chruszcz, M. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
5KRV
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Vibrio vulnificus in complex Arginine | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, ARGININE, ... | | Authors: | Ussin, N, Abdulsalam, R.W, Magee, P, Chruszcz, M. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of 1-deoxy-D-xylulose 5-phosphate Reductoisomerase from Vibrio vulnificus.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
8VUB
 
 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
6QY5
 
 | | Pink beam serial crystallography: Lysozyme, 1 us exposure, 4448 patterns merged (1 chip) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Lieske, J, Tolstikova, A, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
8VUM
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5e2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-6,7-dimethoxyquinazolin-2-yl]amino}piperidin-1-yl)methyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
5NEA
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 4-(2-methyl-1,3-oxazol-5-yl)benzene-1-sulfonammide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-1,3-oxazol-5-yl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Ferraroni, M, Supuran, C.T, Krasavin, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1,3-Oxazole-based selective picomolar inhibitors of cytosolic human carbonic anhydrase II alleviate ocular hypertension in rabbits: Potency is supported by X-ray crystallography of two leads.
Bioorg. Med. Chem., 25, 2017
|
|
6MUG
 
 | |
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MU8
 
 | |
3UET
 
 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
1VTO
 
 | |
4PQU
 
 | | Crystal structure of HIV-1 Reverse Transcriptase in complex with RNA/DNA and dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', ... | | Authors: | Das, K, Bandwar, R.P, Arnold, E. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
4IFY
 
 | | HIV-1 reverse transcriptase with bound fragment at the Knuckles site | | Descriptor: | 1-[4-(trifluoromethoxy)phenyl]methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4ID5
 
 | | HIV-1 reverse transcriptase with bound fragment at the RNase H primer grip site | | Descriptor: | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
1S9A
 
 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
3QO9
 
 | | Crystal structure of HIV-1 Reverse Transcriptase (RT) in complex with TSAO-T, a non-nucleoside RT inhibitor (NNRTI) | | Descriptor: | 1-[(5R,6R,8R,9R)-4-amino-9-{[tert-butyl(dimethyl)silyl]oxy}-6-({[tert-butyl(dimethyl)silyl]oxy}methyl)-2,2-dioxido-1,7-dioxa-2-thiaspiro[4.4]non-3-en-8-yl]-5-methylpyrimidine-2,4(1H,3H)-dione, Reverse transcriptase/ ribonuclease H, p51 RT | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of tert-Butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in Complex with HIV-1 Reverse Transcriptase (RT) Redefines the Elastic Limits of the Non-nucleoside Inhibitor-Binding Pocket.
J.Med.Chem., 54, 2011
|
|
3BGR
 
 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
