2QU6
 
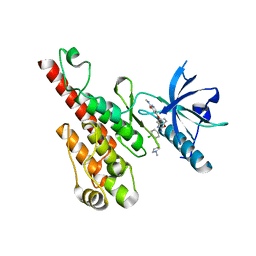 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzoxazole inhibitor | | Descriptor: | 4-({2-[(4-chloro-3-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}phenyl)amino]-1,3-benzoxazol-5-yl}oxy)-N-methylpyridine-2-carboxamide, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Kim, J.L, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2007-08-03 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Evaluation of Orally Active Benzimidazoles and Benzoxazoles as Vascular Endothelial Growth Factor-2 Receptor Tyrosine Kinase Inhibitors.
J.Med.Chem., 50, 2007
|
|
2QU7
 
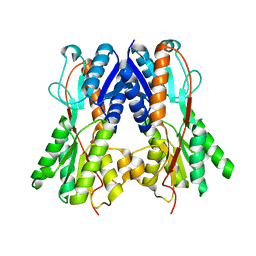 | | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative transcription regulator from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
2QU8
 
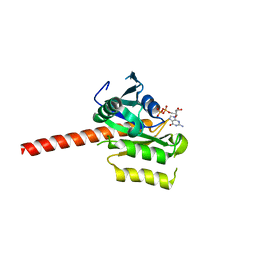 | | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative nucleolar GTP-binding protein 1 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Kozieradzki, I, Zhao, Y, Ravichandran, M, Shapiro, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Sukumar, D, Hassanali, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum.
To be Published
|
|
2QU9
 
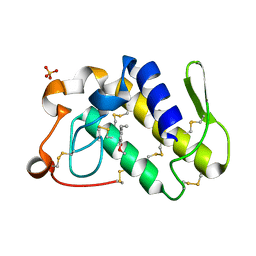 | | Crystal structure of the complex of group II phospholipase A2 with Eugenol | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Vikram, G, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of the complex of group II phospholipase A2 with Eugenol
To be Published
|
|
2QUA
 
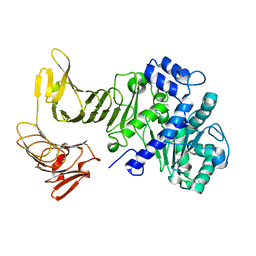 | | Crystal structure of LipA from Serratia marcescens | | Descriptor: | CALCIUM ION, Extracellular lipase | | Authors: | Meier, R, Baumann, U. | | Deposit date: | 2007-08-04 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A calcium-gated lid and a large beta-roll sandwich are revealed by the crystal structure of extracellular lipase from Serratia marcescens.
J.Biol.Chem., 282, 2007
|
|
2QUB
 
 | |
2QUD
 
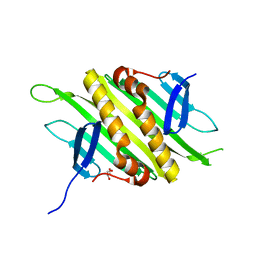 | | PP7 Coat Protein Dimer | | Descriptor: | Coat protein, GLYCEROL | | Authors: | Chao, J.A. | | Deposit date: | 2007-08-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the coevolution of a viral RNA-protein complex.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2QUE
 
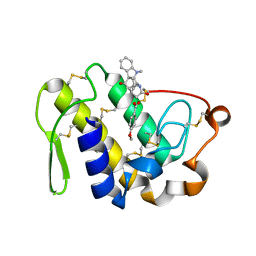 | | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-05 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution
To be Published
|
|
2QUF
 
 | | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus | | Descriptor: | GLYCEROL, Transcription Factor PF0095 | | Authors: | Yang, H, Lipscomb, G.L, Scott, R.A, Rose, J.P, Wang, B.C. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Transcription Factor AXXA-PF0095 from Pyrococcus furiosus
To be Published
|
|
2QUG
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form A | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Hansen, G, Morton, C.J, Pearce, M.C, Feil, S.C, Adams, J.J, Parker, M.W, Bottomley, S.P. | | Deposit date: | 2007-08-05 | | Release date: | 2008-08-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
2QUH
 
 | |
2QUI
 
 | |
2QUJ
 
 | |
2QUK
 
 | |
2QUL
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii at 1.79 A resolution | | Descriptor: | D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUM
 
 | | Crystal structure of D-tagatose 3-epimerase from Pseudomonas cichorii with D-tagatose | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUN
 
 | | Crystal Structure of D-tagatose 3-epimerase from Pseudomonas cichorii in Complex with D-fructose | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamada, M, Nishitani, T, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of D-tagatose 3-epimerase from Pseudomonas cichorii and its complexes with D-tagatose and D-fructose
J.Mol.Biol., 374, 2007
|
|
2QUO
 
 | |
2QUP
 
 | | Crystal structure of uncharacterized protein BH1478 from Bacillus halodurans | | Descriptor: | BH1478 protein, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Rutter, M, Mckenzie, C, Bain, K.T, Smith, D, Ozyurt, S, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-06 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Uncharacterized Protein Bh1478 from Bacillus Halodurans.
To be Published
|
|
2QUQ
 
 | |
2QUR
 
 | | Crystal Structure of F327A/K285P Mutant of cAMP-dependent Protein Kinase | | Descriptor: | 20-mer fragment from cAMP-dependent protein kinase inhibitor alpha, ADENOSINE-5'-DIPHOSPHATE, cAMP-dependent protein kinase, ... | | Authors: | Taylor, S.S, Yang, J, Wu, J. | | Deposit date: | 2007-08-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of non-catalytic core residues to activity and regulation in protein kinase A.
J.Biol.Chem., 284, 2009
|
|
2QUS
 
 | |
2QUT
 
 | |
2QUU
 
 | |
2QUV
 
 | |
