5L3J
 
 | | ESCHERICHIA COLI DNA GYRASE B IN COMPLEX WITH BENZOTHIAZOLE-BASED INHIBITOR | | Descriptor: | 2-[[2-[[4,5-bis(bromanyl)-1~{H}-pyrrol-2-yl]carbonylamino]-1,3-benzothiazol-6-yl]amino]-2-oxidanylidene-ethanoic acid, DNA gyrase subunit B, IODIDE ION | | Authors: | Gjorgjieva, M, Tomasic, T, Barancokova, M, Katsamakas, S, Ilas, J, Montalvao, S, Tammella, P, Peterlin Masic, L, Kikelj, D. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Benzothiazole Scaffold-Based DNA Gyrase B Inhibitors.
J.Med.Chem., 59, 2016
|
|
1AIO
 
 | | CRYSTAL STRUCTURE OF A DOUBLE-STRANDED DNA CONTAINING THE MAJOR ADDUCT OF THE ANTICANCER DRUG CISPLATIN | | Descriptor: | Cisplatin, DNA (5'-D(*CP*CP*(BRU)P*CP*TP*[PT(NH3)2(GP*GP)]*TP*CP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3') | | Authors: | Takahara, P.M, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 1997-04-23 | | Release date: | 1997-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin.
Nature, 377, 1995
|
|
8B4C
 
 | | ToxR bacterial transcriptional regulator bound to 20 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (20-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4E
 
 | | ToxR bacterial transcriptional regulator bound to 25 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (25-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4B
 
 | | ToxR bacterial transcriptional regulator bound to 19 bp ompU promoter DNA | | Descriptor: | AMMONIUM ION, CADMIUM ION, Cholera toxin transcriptional activator, ... | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8B4D
 
 | | ToxR bacterial transcriptional regulator bound to 40 bp toxT promoter DNA | | Descriptor: | Cholera toxin transcriptional activator, DNA (40-MER) | | Authors: | Canals, A, Pieretti, S, Muriel, M, El Yaman, N, Fabrega-Ferrer, M, Perez-Luque, R, Krukonis, E.S, Coll, M. | | Deposit date: | 2022-09-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | ToxR activates the Vibrio cholerae virulence genes by tethering DNA to the membrane through versatile binding to multiple sites.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1NUI
 
 | | Crystal Structure of the primase fragment of Bacteriophage T7 primase-helicase protein | | Descriptor: | DNA primase/helicase, MAGNESIUM ION, ZINC ION | | Authors: | Kato, M, Ito, T, Wagner, G, Richardson, C.C, Ellenberger, T. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modular Architecture of the Bacteriophage T7 Primase Couples RNA primer Synthesis to DNA Synthesis
Mol.Cell, 11, 2003
|
|
235D
 
 | |
234D
 
 | |
1PIK
 
 | | ESPERAMICIN A1-DNA COMPLEX, NMR, 4 STRUCTURES | | Descriptor: | 2,4-dideoxy-3-O-methyl-4-(propan-2-ylamino)-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-S-methyl-4-thio-beta-D-ribo-hexopyranose, 2-deoxy-alpha-L-fucopyranose, ... | | Authors: | Kumar, R.A, Ikemoto, N, Patel, D.J. | | Deposit date: | 1996-12-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Esperamicin A1-DNA Complex
J.Mol.Biol., 265, 1997
|
|
4DQY
 
 | |
236D
 
 | |
215D
 
 | |
3FHG
 
 | |
3FHF
 
 | |
292D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE:THE CRYSTAL STRUCTURE OF THE D(CG)3 AND N-(2-AMINOETHYL)-1,4-DIAMINOBUTANE COMPLEX | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ohishi, H, Kunisawa, S, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Tomita, K, Hakoshima, T. | | Deposit date: | 1991-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine. The crystal structure of the d(CG)3 and N-(2-aminoethyl)-1,4-diamino-butane complex.
FEBS Lett., 284, 1991
|
|
7XF1
 
 | | Crystal strucutre of apoCasDinG in complex with ssDNA | | Descriptor: | CasDinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3') | | Authors: | Zhang, J.T, Cui, N, Liu, Y.R, Huang, H.D, Jia, N. | | Deposit date: | 2022-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Type IV-A CRISPR-Csf complex: Assembly, dsDNA targeting, and CasDinG recruitment.
Mol.Cell, 83, 2023
|
|
1D36
 
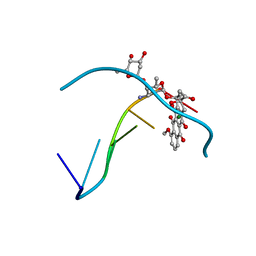 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4BAC
 
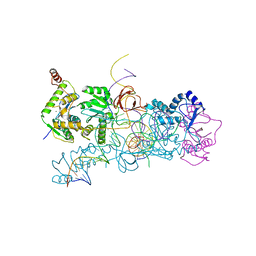 | | prototype foamy virus strand transfer complexes on product DNA | | Descriptor: | 5'-D(*AP*GP*GP*AP*GP*CP*CP*AP*AP*GP*AP*CP*GP*GP *AP*TP*CP)-3', 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*TP*GP*TP*AP)-3', DNA (38-MER), ... | | Authors: | Yin, Z, Lapkouski, M, Yang, W, Craigie, R. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Protein Sci., 21, 2012
|
|
1D56
 
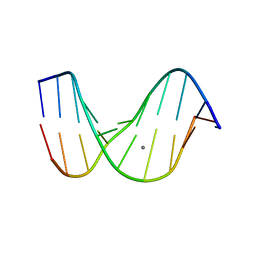 | |
1D35
 
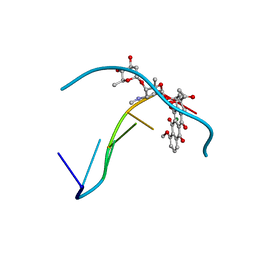 | | FACILE FORMATION OF A CROSSLINKED ADDUCT BETWEEN DNA AND THE DAUNORUBICIN DERIVATIVE MAR70 MEDIATED BY FORMALDEHYDE: MOLECULAR STRUCTURE OF THE MAR70-D(CGTNACG) COVALENT ADDUC | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, DNA (5'-D(*CP*GP*TP*(A40)P*CP*G)-3'), MAGNESIUM ION | | Authors: | Gao, Y.-G, Liaw, Y.-C, Li, Y.-K, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Facile formation of a crosslinked adduct between DNA and the daunorubicin derivative MAR70 mediated by formaldehyde: molecular structure of the MAR70-d(CGTnACG) covalent adduct.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ODG
 
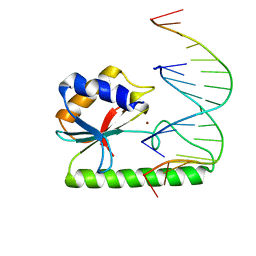 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
6WRB
 
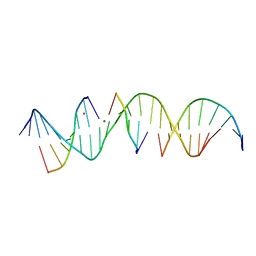 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J5 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*CP*GP*AP*CP*GP*GP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WQG
 
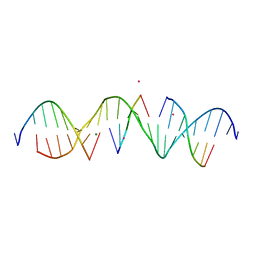 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J3 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-28 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6WRI
 
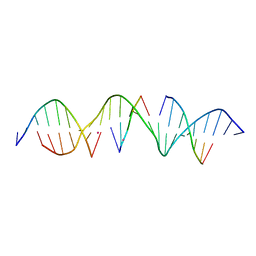 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J28 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*TP*GP*AP*CP*TP*CP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-04-29 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.057 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
