7A7H
 
 | | Crystal structure of PPARgamma in complex with compound TK90 | | Descriptor: | (2~{R})-2-[[4-[[4-methoxy-2-(trifluoromethyl)phenyl]methylcarbamoyl]phenyl]methyl]butanoic acid, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Ni, X, Kirchner, T, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combined Cardioprotective and Adipocyte Browning Effects Promoted by the Eutomer of Dual sEH/PPAR gamma Modulator.
J.Med.Chem., 64, 2021
|
|
5LKV
 
 | |
6ZW2
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
5LLC
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 3-(Methylamino)-2,5,6-trifluoro-4-[(2-phenylethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,6-tris(fluoranyl)-5-(methylamino)-4-(2-phenylethylsulfonyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
5LM6
 
 | | VIM-2 metallo-beta-lactamase in complex with 2-(3-fluoro-4-hydroxyphenyl)-3-oxoisoindoline-4-carboxylic acid (compound 35) | | Descriptor: | 2-(3-fluoranyl-4-oxidanyl-phenyl)-3-oxidanylidene-1~{H}-isoindole-4-carboxylic acid, FORMIC ACID, Metallo-beta-lactamase VIM-2, ... | | Authors: | Li, G.-B, Brem, J, Someya, H, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | NMR-filtered virtual screening leads to non-metal chelating metallo-beta-lactamase inhibitors.
Chem Sci, 8, 2017
|
|
6SYN
 
 | | Crystal structure of Y. pestis penicillin-binding protein 3 | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Pankov, G, Hunter, W.N, Dawson, A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The structure of penicillin-binding protein 3 from Yersinia pestis
To Be Published
|
|
5LMF
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator bound to Dcp2 HLM3 peptide (region 484-500) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Charenton, C, Gaudon-Plesse, C, Fourati, Z, Taverniti, V, Back, R, Kolesnikova, O, Seraphin, B, Graille, M. | | Deposit date: | 2016-07-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique surface on Pat1 C-terminal domain directly interacts with Dcp2 decapping enzyme and Xrn1 5'-3' mRNA exonuclease in yeast.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6SYS
 
 | | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2 | | Descriptor: | (4~{S})-4-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, GLYCEROL, ZINC ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-10-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of (3aR,4S,9bS)-4-(4-hydroxyphenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide with carbonic anhydrase 2
To Be Published
|
|
6ZXI
 
 | | Crystal Structure of the OXA-48 Carbapenem-Hydrolyzing Class D beta-Lactamase in Complex with the DBO inhibitor ANT3310 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
J.Med.Chem., 63, 2020
|
|
6SVK
 
 | | human Myeloid-derived growth factor (MYDGF) | | Descriptor: | MALONATE ION, Myeloid-derived growth factor | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure and receptor-interacting residues of MYDGF - a protein mediating ischemic tissue repair.
Nat Commun, 10, 2019
|
|
5LO7
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
6SVQ
 
 | | Crystal structure of human GFAT-1 G461E after UDP-GlcNAc soaking | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate-aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SWK
 
 | | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus | | Descriptor: | Histidine kinase | | Authors: | Lansky, S, Shiradsky, M, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | The kinase domain of GanS, a histidine kinase from Geobacillus stearothermophilus
To Be Published
|
|
6ZYU
 
 | | Structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM549 | | Descriptor: | (4~{R})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, (4~{S})-4-cyclopropyl-7-fluoranyl-3,4-dihydro-2~{H}-thiochromene 1,1-dioxide, ACETATE ION, ... | | Authors: | Dorosz, J, Christensen, K.M, Kastrup, J.S. | | Deposit date: | 2020-08-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Thiochroman Dioxide Analogues of Benzothiadiazine Dioxides as New Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors.
Acs Chem Neurosci, 12, 2021
|
|
6SZ5
 
 | |
5M2A
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
7A36
 
 | |
6SX9
 
 | | Crystal structure of carbonic anhydrase 2 with 4-(2-chlorophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide | | Descriptor: | 4-(2-chlorophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2019-09-25 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of carbonic anhydrase 2 with 4-(2-chlorophenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-sulfonamide
To Be Published
|
|
6SZH
 
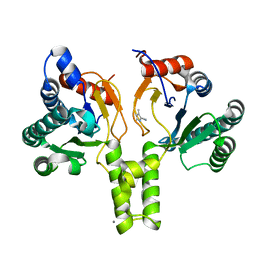 | | Acinetobacter baumannii undecaprenyl pyrophosphate synthase (AB-UppS) in complex with GW197 | | Descriptor: | 3,5-dimethyl-1~{H}-pyrrole-2-carbonitrile, CALCIUM ION, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-10-02 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cocktailed fragment screening by X-ray crystallography of the antibacterial target undecaprenyl pyrophosphate synthase from Acinetobacter baumannii.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5LPE
 
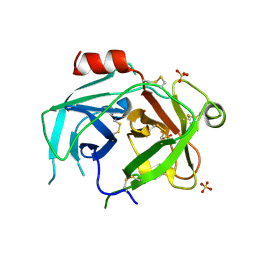 | | Kallikrein-related peptidase 10 complex with Zn2+ | | Descriptor: | Kallikrein-10, SULFATE ION, ZINC ION | | Authors: | Goettig, P, Debela, M, Magdolen, V, Bode, W, Brandstetter, H. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10.
Biol.Chem., 397, 2016
|
|
6SXF
 
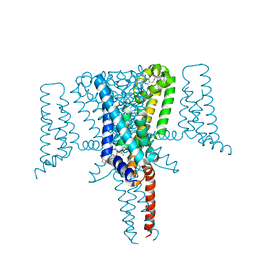 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Tamoxifen (2.8 Angstrom resolution) | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
7A3W
 
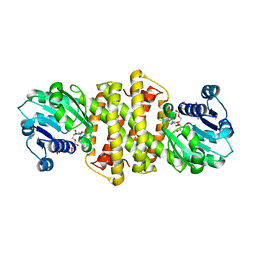 | | Structure of Imine Reductase from Pseudomonas sp. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NAD(P)-dependent oxidoreductase, ... | | Authors: | Cuetos, A, Thorpe, T, Turner, N.J, Grogan, G. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Multifunctional biocatalyst for conjugate reduction and reductive amination.
Nature, 604, 2022
|
|
7A39
 
 | |
6SZW
 
 | |
7A3E
 
 | |
