7APH
 
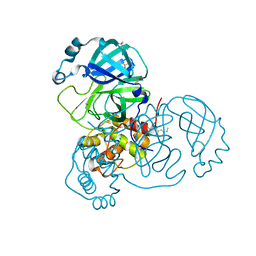 | | Structure of SARS-CoV-2 Main Protease bound to Tofogliflozin. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-16 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7RDE
 
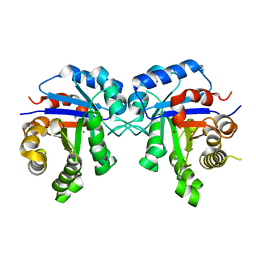 | | Human Triose Phosphate Isomerase Q181P | | Descriptor: | BROMIDE ION, CALCIUM ION, Triosephosphate isomerase | | Authors: | VanDemark, A.P, Kowalski, J. | | Deposit date: | 2021-07-09 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Itavastatin and resveratrol increase triosephosphate isomerase protein in a newly identified variant of TPI deficiency.
Dis Model Mech, 15, 2022
|
|
1H2Z
 
 | |
5Z65
 
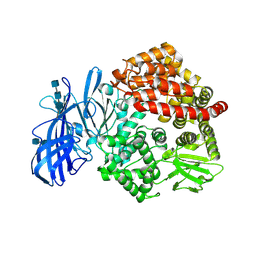 | | Crystal structure of porcine aminopeptidase N ectodomain in functional form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, Y.G, Li, R, Jiang, L.G, Qiao, S.L, Zhang, G.P. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization of the interaction between recombinant porcine aminopeptidase N and spike glycoprotein of porcine epidemic diarrhea virus.
Int. J. Biol. Macromol., 117, 2018
|
|
6Z1J
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV LSP co-crystallization with spheroidene | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A.G, Fufina, T.Y, Vasilieva, L.G, Betzel, C, Selikhanov, G.K. | | Deposit date: | 2020-05-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel approaches for the lipid sponge phase crystallization of the Rhodobacter sphaeroides photosynthetic reaction center.
Iucrj, 7, 2020
|
|
5ZEF
 
 | | Crystal structure of Entamoeba histolytica Arginase in complex with L- Norvaline at 2.01 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, GLYCEROL, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
6W9T
 
 | |
6Y3O
 
 | | 14-3-3 Sigma in complex with phosphorylated CAMKK2 peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CAMKK2, ... | | Authors: | Ballone, A, Lau, R.A, Zweipfenning, F.P.A, Ottmann, C. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new soaking procedure for X-ray crystallographic structural determination of protein-peptide complexes.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6W90
 
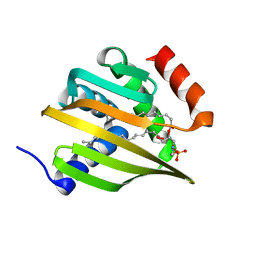 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
7RAN
 
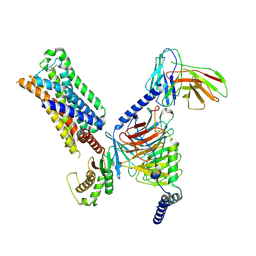 | | 5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (3R)-3-methyl-5-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,3,6-tetrahydropyridin-1-ium, 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Nature, 610, 2022
|
|
6VSB
 
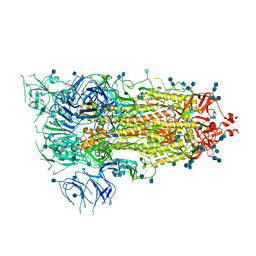 | | Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Wang, N, Corbett, K.S, Goldsmith, J.A, Hsieh, C, Abiona, O, Graham, B.S, McLellan, J.S. | | Deposit date: | 2020-02-10 | | Release date: | 2020-02-26 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.
Science, 367, 2020
|
|
6YAP
 
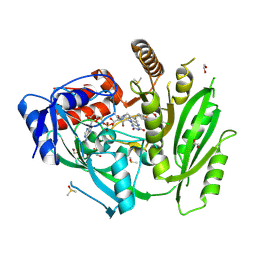 | | Crystal structure of ZmCKO4a in complex with inhibitor 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-Chloro-5-trifluoromethoxy-phenyl)-3-[2-(2-hydroxy-ethyl)-phenyl]-urea, Cytokinin dehydrogenase 4, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diphenylurea-derived cytokinin oxidase/dehydrogenase inhibitors for biotechnology and agriculture.
J.Exp.Bot., 72, 2021
|
|
7R6J
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 1 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[3-methyl-5-(pyrimidin-2-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
6Y8Z
 
 | | Structure of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (PGM5) | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Gustafsson, R, Eckhard, U, Selmer, M. | | Deposit date: | 2020-03-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Characterization of Phosphoglucomutase 5 from Atlantic and Baltic Herring-An Inactive Enzyme with Intact Substrate Binding.
Biomolecules, 10, 2020
|
|
7R5C
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7R1G
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-38 (R207C, D210S, S211V) | | Descriptor: | Beta-aspartyl-peptidase, Isoaspartyl peptidase, SODIUM ION | | Authors: | Loch, J.I, Kadziolka, K, Jaskolski, M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6ZDX
 
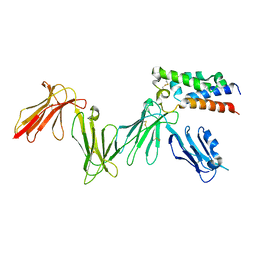 | | RIFIN variable region bound to LILRB1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leukocyte immunoglobulin-like receptor subfamily B member 1, ... | | Authors: | Harrison, T.E, Higgins, M.K. | | Deposit date: | 2020-06-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RIFIN-mediated activation of LILRB1 in malaria.
Nature, 587, 2020
|
|
7ROA
 
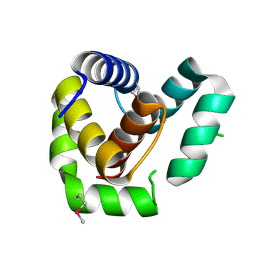 | | Crystal structure of EntV136 from Enterococcus faecalis | | Descriptor: | EntV | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-07-30 | | Release date: | 2022-10-12 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
7RKT
 
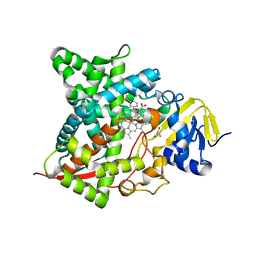 | |
8GN5
 
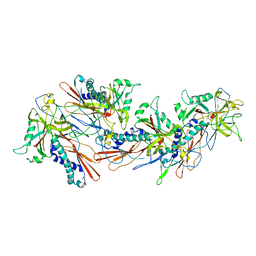 | | Designed pH-responsive P22 VLP | | Descriptor: | Major capsid protein | | Authors: | Kim, K.J, Kim, G, Bae, J.H, Song, J.J, Kim, H.S. | | Deposit date: | 2022-08-23 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A pH-Responsive Virus-Like Particle as a Protein Cage for a Targeted Delivery.
Adv Healthc Mater, 13, 2024
|
|
7RKR
 
 | |
7RKW
 
 | |
7REV
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 3 | | Descriptor: | (2R,3R,4R,5S)-1-[(4-{[4-(furan-2-yl)-2-methylanilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-13 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5ZEH
 
 | | Crystal structure of Entamoeba histolytica Arginase in complex with L- Ornithine at 2.35 A | | Descriptor: | 1,2-ETHANEDIOL, Arginase, L-ornithine, ... | | Authors: | Malik, A, Dalal, V, Ankri, S, Tomar, S. | | Deposit date: | 2018-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural insights into Entamoeba histolytica arginase and structure-based identification of novel non-amino acid based inhibitors as potential antiamoebic molecules.
Febs J., 286, 2019
|
|
7C0P
 
 | |
