6Q1Q
 
 | |
6FQH
 
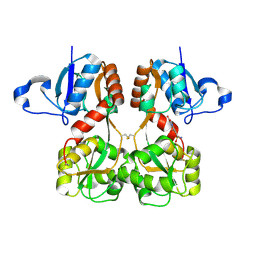 | | GluA2(flop) S729C ligand binding core dimer bound to NBQX at 1.76 Angstrom resolution | | Descriptor: | 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75940013 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
9F15
 
 | | Carbonic anhydrase II variant with bound iron complex | | Descriptor: | 4,4,7,10,10-pentamethyl-3,6,8,11-tetrakis(oxidanylidene)-N-[2-(4-sulfamoylphenyl)ethyl]-2,5,7,9,12-pentazabicyclo[11.4.0]heptadeca-1(13),14,16-triene-15-carboxamide containing iron, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Directed Evolution of an Artificial Hydroxylase Based on a Thermostable Human Carbonic Anhydrase Protein
Acs Catalysis, 14, 2024
|
|
7OC2
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2295 | | Descriptor: | Cyclic 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE-(7-3)-7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE-(7-19)-N-ACETYL-L-CYSTEINE-(8-25)-[3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID-()-(6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-04-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
8HLT
 
 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
5JDQ
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 100 mM Na+ and 10mM Sr2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8OEG
 
 | | PDE4B bound to MAPI compound 92a | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1~{S})-2-[3,5-bis(chloranyl)-1-oxidanyl-pyridin-4-yl]-1-(3,4-dimethoxyphenyl)ethyl] 5-[[[(1~{R})-2-[[(3~{R})-1-azabicyclo[2.2.2]octan-3-yl]oxy]-2-oxidanylidene-1-phenyl-ethyl]amino]methyl]thiophene-2-carboxylate, ... | | Authors: | Rizzi, A, Armani, E. | | Deposit date: | 2023-03-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of M 3 Antagonist-PDE4 Inhibitor (MAPI) Dual Pharmacology Molecules for the Treatment of COPD.
J.Med.Chem., 66, 2023
|
|
9C34
 
 | | Proline utilization A with the FADH- N5 atom covalently modified by proline | | Descriptor: | (2S)-3,4-dihydro-2H-pyrrole-2-carboxylic acid, (2~{S},5~{R})-5-[10-[(2~{S},3~{S},4~{R})-5-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2,3,4-tris(oxidanyl)pentyl]-7,8-dimethyl-2,4-bis(oxidanylidene)benzo[g]pteridin-5-yl]pyrrolidine-2-carboxylic acid, Bifunctional protein PutA, ... | | Authors: | Tanner, J.J, Buckley, D.P. | | Deposit date: | 2024-05-31 | | Release date: | 2025-06-04 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Visualization of covalent intermediates and conformational states of proline utilization A by X-ray crystallography and molecular dynamics simulations.
J.Biol.Chem., 301, 2025
|
|
7ZPR
 
 | | KtrAB complex with N-terminal deletion of KtrB 1-19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Vonck, J, Stautz, J. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | KtrAB complex with N-terminal deletion of KtrB 1-19
To Be Published
|
|
8V5I
 
 | | Crystal structure of MAP4K4 in complex with an inhibitor | | Descriptor: | (3M)-N~6~-(1,4-dimethyl-1H-pyrazol-3-yl)-3-(1-methyl-1H-imidazol-5-yl)-2,7-naphthyridine-1,6-diamine, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Greasley, S.E, Diehl, W. | | Deposit date: | 2023-11-30 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Microtubule-Associated Serine/Threonine Kinase-like (MASTL).
J.Med.Chem., 67, 2024
|
|
7ZI1
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the dCKi1 inhibitor | | Descriptor: | Deoxycytidine kinase, N-[3-[[4-(4-azanylpyrimidin-2-yl)-1,3-thiazol-2-yl]amino]-4-methyl-phenyl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
9C35
 
 | |
6SSO
 
 | | EDN mutant L45H | | Descriptor: | ACETATE ION, Non-secretory ribonuclease | | Authors: | Fernandez-Millan, P, Prats-Ejarque, G, Vazquez-Monteagudo, S, Boix, E. | | Deposit date: | 2019-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and functional characterization of new family enzymes derivates from human RNase 1 and 3 with antimicrobial and ribonuclease activity
To Be Published
|
|
7ZI5
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0274 inhibitor | | Descriptor: | 4-(4-azanylpyrimidin-2-yl)-N-[2-methyl-5-[4-[(4-methylpiperazin-1-yl)methyl]phenyl]phenyl]-1,3-thiazol-2-amine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
7ZI2
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the dCKi2 inhibitor | | Descriptor: | Deoxycytidine kinase, N-[3-[[4-[4,6-bis(azanyl)pyrimidin-2-yl]-1,3-thiazol-2-yl]amino]-4-methyl-phenyl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
8EWV
 
 | | DNA-encoded library (DEL)-enabled discovery of proximity inducing small molecules | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Schreiber, S.L, Shu, W, Ma, X, Michaud, G, Bonazzi, S, Berst, F. | | Deposit date: | 2022-10-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | DNA-encoded library-enabled discovery of proximity-inducing small molecules.
Nat.Chem.Biol., 20, 2024
|
|
6D6M
 
 | |
7ZIA
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0634 inhibitor | | Descriptor: | 2-[2-[[2-methyl-5-[6-(4-methylpiperazin-1-yl)sulfonylpyridin-3-yl]phenyl]-propyl-amino]-1,3-thiazol-4-yl]pyrimidine-4,6-diamine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Ben-Yaala, K, Saez-Ayala, M, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
9JBH
 
 | | Cryo-EM structure of the human LYCHOS PLD homodimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, Lysosomal cholesterol signaling protein | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
5CI5
 
 | | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose | | Descriptor: | 1,2-ETHANEDIOL, Extracellular solute-binding protein family 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of an ABC transporter Solute Binding Protein from Thermotoga Lettingae TMO (Tlet_1705, TARGET EFI-510544) bound with alpha-D-Tagatose
To be published
|
|
4R3J
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin | | Descriptor: | (2R)-2-[(1R)-1-(acetylamino)-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin
To be Published
|
|
7ZI9
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0624 inhibitor | | Descriptor: | 2-[2-[[2-methyl-5-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]-propyl-amino]-1,3-thiazol-4-yl]pyrimidine-4,6-diamine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
6Y80
 
 | | Fragment KCL_916 in complex with MAP kinase p38-alpha | | Descriptor: | 1-(2-adamantylmethyl)-3-ethyl-guanidine, 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, CALCIUM ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-02 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
9JBG
 
 | | Cryo-EM structure of the human LYCHOS in complex with lipids in the expanded state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Lysosomal cholesterol signaling protein, ... | | Authors: | Yu, S, Liang, L. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-16 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights into cholesterol sensing by the LYCHOS-mTORC1 pathway.
Nat Commun, 16, 2025
|
|
4R53
 
 | |
