6VN4
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 1 | | Descriptor: | 3-({4-hydroxy-1-[(2R)-2-methyl-3-phenylpropanoyl]piperidin-4-yl}methyl)quinazolin-4(3H)-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6VO8
 
 | | X-ray structure of the Cj1427 in the presence of NADH and GDP-D-glycero-D-mannoheptose, an essential NAD-dependent dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative sugar-nucleotide epimerase/dehydratease, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{S},5~{S},6~{S})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Spencer, K.D, Anderson, T.K, Thoden, J.B, Huddleston, J.P, Raushel, F.M, Holden, H.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Cj1427, an Essential NAD-Dependent Dehydrogenase for the Biosynthesis of the Heptose Residues in the Capsular Polysaccharides ofCampylobacter jejuni.
Biochemistry, 59, 2020
|
|
6VR4
 
 | |
6VUW
 
 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT368 | | Descriptor: | (7R)-7-methyl-2-({[(3R)-1-methylpiperidin-3-yl]methyl}sulfanyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4-amine, GLYCEROL, N-acetyltransferase Eis, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VV6
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB113 | | Descriptor: | 1-(4-fluorophenyl)-5-[3-(1H-indol-3-yl)propoxy]-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B, Chaves-Pacheco, S.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VVW
 
 | |
6W04
 
 | |
6W2Q
 
 | | Junction 34, DHR53-DHR4 | | Descriptor: | CALCIUM ION, Junction 34 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3S
 
 | | Crystal structure of ligand-binding domain of Campylobacter jejuni chemoreceptor Tlp3 in complex with L-leucine | | Descriptor: | GLYCEROL, LEUCINE, Methyl-accepting chemotaxis protein, ... | | Authors: | Khan, M.F, Machuca, M.A, Rahman, M.M, Roujeinikova, A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by theCampylobacter jejuniChemoreceptor Tlp3.
Biomolecules, 10, 2020
|
|
7ZVA
 
 | | Crystal Structure of the native zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 1.80 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C-terminal peptidase, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZU8
 
 | | Crystal Structure of the zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 2.05 A resolution in presence of the crystallophore Lu-Xo4. | | Descriptor: | 12-oxidanyl-9,11$l^{3}-dioxa-1$l^{4},19$l^{4},22,27$l^{4},28$l^{4}-pentaza-10$l^{6}-lutetaoctacyclo[17.5.2.1^{3,7}.1^{10,13}.0^{1,10}.0^{10,19}.0^{10,28}.0^{17,27}]octacosa-3,5,7(28),11,13,15,17(27)-heptaen-8-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
1K5D
 
 | | Crystal structure of Ran-GPPNHP-RanBP1-RanGAP complex | | Descriptor: | GTP-binding nuclear protein RAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1K5S
 
 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PPA | | Descriptor: | CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, PENICILLIN G ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K6G
 
 | | Solution Structure of Conserved AGNN Tetraloops: Insights into Rnt1p RNA Processing | | Descriptor: | RNA (5'-R(P*GP*GP*CP*GP*UP*CP*AP*UP*GP*AP*GP*UP*CP*CP*AP*UP*GP*GP*CP*GP*CP*C)-3') | | Authors: | Lebars, I, Lamontagne, B, Yoshizawa, S, Abou Elela, S, Fourmy, D. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing.
EMBO J., 20, 2001
|
|
1K70
 
 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K7G
 
 | | PrtC from Erwinia chrysanthemi | | Descriptor: | CALCIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Hege, T, Baumann, U. | | Deposit date: | 2001-10-19 | | Release date: | 2002-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protease C of Erwinia chrysanthemi: The Crystal Structure and Role of Amino Acids Y228 and E189
J.Mol.Biol., 314, 2001
|
|
1K88
 
 | | Crystal structure of procaspase-7 | | Descriptor: | procaspase-7 | | Authors: | Chai, J, Wu, Q, Shiozaki, E, Srinivasa, S.M, Alnemri, E.S, Shi, Y. | | Deposit date: | 2001-10-23 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding
Cell(Cambridge,Mass.), 107, 2001
|
|
6ZRH
 
 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
1K81
 
 | |
1K8G
 
 | |
6ZSX
 
 | |
6ZTC
 
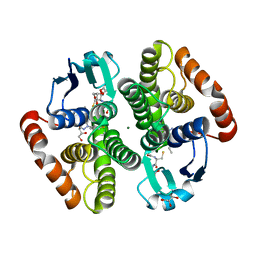 | | CRYSTAL STRUCTURE OF PROSTAGLANDIN D2 SYNTHASE IN COMPLEX WITH FRAGMENT 1A AT 1.84A RESOLUTION. | | Descriptor: | 1-[1-(3-fluorophenyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]propan-1-one, GLUTATHIONE, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
1K8Y
 
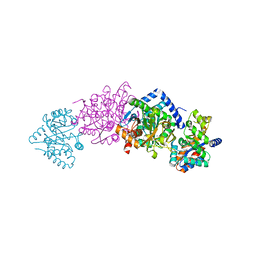 | | CRYSTAL STRUCTURE OF THE TRYPTOPHAN SYNTHASE BETA-SER178PRO MUTANT COMPLEXED WITH D,L-ALPHA-GLYCEROL-3-PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-26 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
