7WPP
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 2.85 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPS
 
 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (7 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPR
 
 | | VWF D'D3 dimer complexed with D1D2 at 4.39 angstron resolution(VWF tube) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
8JCH
 
 | |
8JRC
 
 | |
8HZR
 
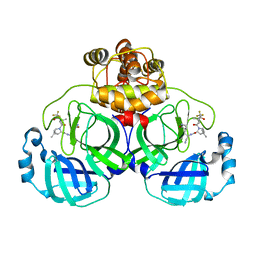 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2023-01-09 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with PF07321332
To Be Published
|
|
8K5P
 
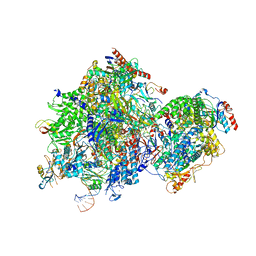 | |
7VBD
 
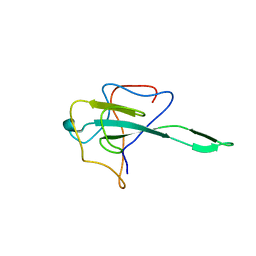 | | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0 | | Descriptor: | Nucleoprotein | | Authors: | Zeng, P, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0
To Be Published
|
|
3CL5
 
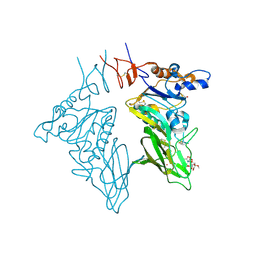 | | Structure of coronavirus hemagglutinin-esterase in complex with 4,9-O-diacetyl sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CL4
 
 | | Crystal structure of bovine coronavirus hemagglutinin-esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Zeng, Q.H, Langereis, M.A, van Vliet, A.L.W, Huizinga, E.G, de Groot, R.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7W7R
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7W7S
 
 | | High resolution structure of a fish aquaporin reveals a novel extracellular fold. | | Descriptor: | Aquaporin 1 | | Authors: | Zeng, J, Schmitz, F, Isaksson, S, Glas, J, Arbab, O, Andersson, M, Sundell, K, Eriksson, L, Swaminathan, K, Tornroth-Horsefield, S, Hedfalk, K. | | Deposit date: | 2021-12-06 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution structure of a fish aquaporin reveals a novel extracellular fold.
Life Sci Alliance, 5, 2022
|
|
7YBG
 
 | |
5U4I
 
 | | Structural Basis of Co-translational Quality Control by ArfA and RF2 Bound to Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zeng, F, Chen, Y, Remis, J, Shekhar, M, Phillips, J.C, Tajkhorshid, E, Jin, H. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-11 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Nature, 541, 2017
|
|
5M1Y
 
 | |
4W55
 
 | | T4 Lysozyme L99A with n-Propylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, propylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6401 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W58
 
 | | T4 Lysozyme L99A with n-Pentylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, pentylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6VNT
 
 | | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution.
To be Published
|
|
7SA4
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tian, Y, Zeng, F, Raybarman, A, Carruthers, A, Li, Q, Fatma, S, Huang, R.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Sequential rescue and repair of stalled and damaged ribosome by bacterial PrfH and RtcB.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8HFO
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7d | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(thiophen-2-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFN
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7b | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(6-methoxypyridin-3-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
4W56
 
 | | T4 Lysozyme L99A with sec-Butylbenzene Bound | | Descriptor: | (2R)-butan-2-ylbenzene, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W57
 
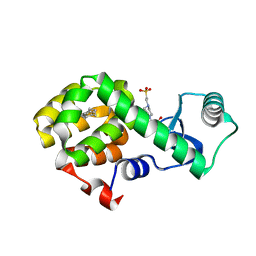 | | T4 Lysozyme L99A with n-Butylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, N-BUTYLBENZENE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6801 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W59
 
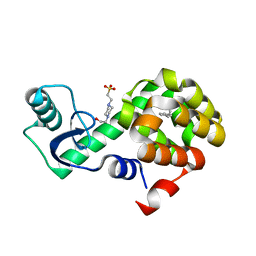 | | T4 Lysozyme L99A with n-Hexylbenzene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, hexylbenzene | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8H5A
 
 | |
