1T4B
 
 | | 1.6 Angstrom structure of Esherichia coli aspartate-semialdehyde dehydrogenase. | | Descriptor: | Aspartate-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
2FBW
 
 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
1XSP
 
 | | Crystal Structure of human DNA polymerase lambda in complex with nicked DNA and pyrophosphate | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Kunkel, T.A, Pedersen, L.C. | | Deposit date: | 2004-10-19 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A closed conformation for the Pol lambda catalytic cycle.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
2GUM
 
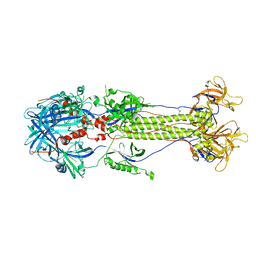 | |
1ZF9
 
 | | GGG Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5PU9
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 187) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PUQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 204) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PV8
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 222) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PVN
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 238) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PW2
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 253) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6YBZ
 
 | | Crystal structure of the D116N mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Gushchin, I, Gordeliy, V. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular mechanism of light-driven sodium pumping.
Nat Commun, 11, 2020
|
|
4XB2
 
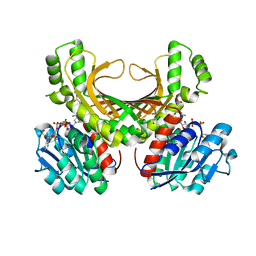 | | Hyperthermophilic archaeal homoserine dehydrogenase mutant in complex with NADPH | | Descriptor: | 319aa long hypothetical homoserine dehydrogenase, L-HOMOSERINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sakuraba, H, Inoue, S, Yoneda, K, Ohshima, T. | | Deposit date: | 2014-12-16 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures of a Hyperthermophilic Archaeal Homoserine Dehydrogenase Suggest a Novel Cofactor Binding Mode for Oxidoreductases.
Sci Rep, 5, 2015
|
|
6PH5
 
 | |
4XDT
 
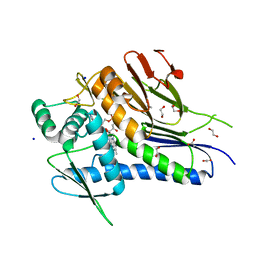 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, a bifunctional FMN transferase/FAD pyrophosphatase, N55Y mutant, FAD bound form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FAD:protein FMN transferase, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2014-12-19 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Evidence for Posttranslational Protein Flavinylation in the Syphilis Spirochete Treponema pallidum: Structural and Biochemical Insights from the Catalytic Core of a Periplasmic Flavin-Trafficking Protein.
Mbio, 6, 2015
|
|
6C89
 
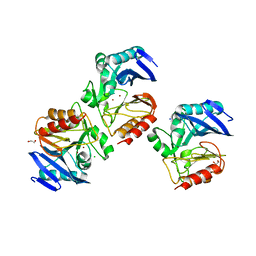 | | NDM-1 Beta-Lactamase Exhibits Differential Active Site Sequence Requirements for the Hydrolysis of Penicillin versus Carbapenem Antibiotics | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Palzkill, T, Sun, Z, Sankaran, B. | | Deposit date: | 2018-01-24 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75006151 Å) | | Cite: | Differential active site requirements for NDM-1 beta-lactamase hydrolysis of carbapenem versus penicillin and cephalosporin antibiotics.
Nat Commun, 9, 2018
|
|
6YHN
 
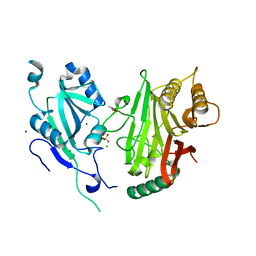 | | Crystal structure of domains 4-5 of CNFy from Yersinia pseudotuberculosis | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, Cytotoxic necrotizing factor, ... | | Authors: | Lukat, P, Gazdag, E.M, Heidler, T.V, Blankenfeldt, W. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bacterial cytotoxic necrotizing factor CNF Y reveals molecular building blocks for intoxication.
Embo J., 40, 2021
|
|
3X3X
 
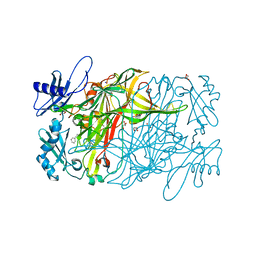 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | Descriptor: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
5PQF
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 52) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PQU
 
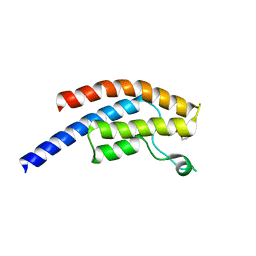 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 67) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
1HN1
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (ORTHORHOMBIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION, SODIUM ION | | Authors: | Juers, D.H, Matthews, B.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Reversible lattice repacking illustrates the temperature dependence of macromolecular interactions.
J.Mol.Biol., 311, 2001
|
|
5PRA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 83) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PRS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 99) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSA
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 118) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PSS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 136) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
