4RQR
 
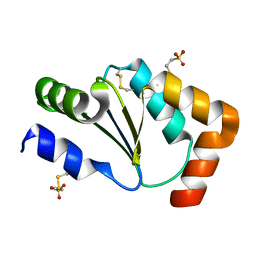 | | Crystal Structure of Human Glutaredoxin with MESNA | | Descriptor: | 1-THIOETHANESULFONIC ACID, Glutaredoxin-1 | | Authors: | Badger, J, Sridhar, V, Logan, C, Hausheer, F.H, Nienaber, V.L. | | Deposit date: | 2014-11-04 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Cysteine Specific Targeting of the Functionally Distinct Peroxiredoxin and Glutaredoxin Proteins by the Investigational Disulfide BNP7787.
Molecules, 20, 2015
|
|
7RJR
 
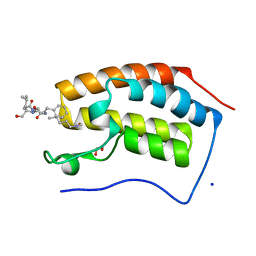 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with BCLTF1 | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-associated transcription factor 1, Bromodomain-containing protein 4, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
7C7M
 
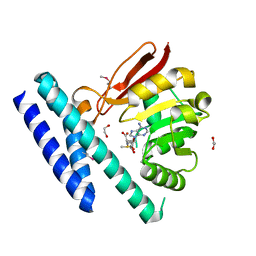 | | The structure of SAM-bound CntL, an aminobutyrate transferase in staphylopine biosysnthesis | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYLMETHIONINE, Staphylopine biosynthesis enzyme CntL | | Authors: | Luo, Z, Luo, S, Zhou, H. | | Deposit date: | 2020-05-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the ligand recognition and catalysis of the key aminobutanoyltransferase CntL in staphylopine biosynthesis.
Faseb J., 35, 2021
|
|
7V1C
 
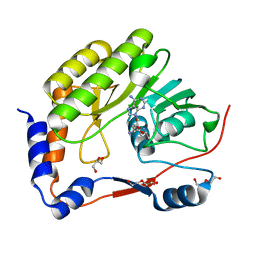 | |
7RJQ
 
 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
7V1E
 
 | |
4ZXI
 
 | | Crystal Structure of holo-AB3403 a four domain nonribosomal peptide synthetase bound to AMP and Glycine | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Drake, E.J, Miller, B.R, Allen, C.L, Gulick, A.M. | | Deposit date: | 2015-05-20 | | Release date: | 2015-12-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of two distinct conformations of holo-non-ribosomal peptide synthetases.
Nature, 529, 2016
|
|
7V1D
 
 | |
8P90
 
 | | TARGET COMPLEX 2 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-26 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAPE-PLD is target of thiazide diuretics.
Cell Chem Biol, 32, 2025
|
|
8U7U
 
 | | Proteasome 20S Core Particle from Beta 3 D205 deletion | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7RJK
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with hnRNPK | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
4ZYQ
 
 | |
5HC0
 
 | | Structure of esterase Est22 with p-nitrophenol | | Descriptor: | ACETIC ACID, GLYCEROL, Lipolytic enzyme, ... | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
8PJE
 
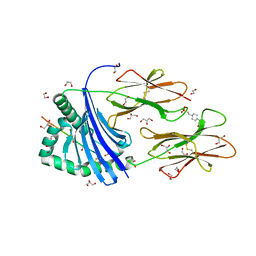 | | Human Leukocyte Antigen class II allotype DR1 presenting influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAT | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Hesketh, S.J, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
7P6N
 
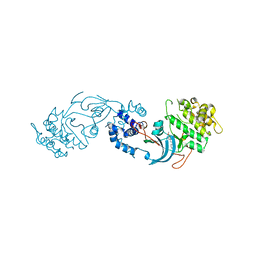 | | ROCK2 IN COMPLEX WITH COMPOUND 12 | | Descriptor: | Rho-associated protein kinase 2, ~{N}-[(1~{R})-1-(3-methoxyphenyl)ethyl]-4-pyridin-4-yl-piperidine-1-carboxamide | | Authors: | Maillard, M.C. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of a Potent, Selective, and Brain-Penetrant Rho Kinase Inhibitor and its Activity in a Mouse Model of Huntington's Disease.
J.Med.Chem., 65, 2022
|
|
8BLV
 
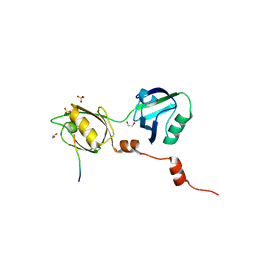 | | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syndecan-4, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide
To Be Published
|
|
6ZBI
 
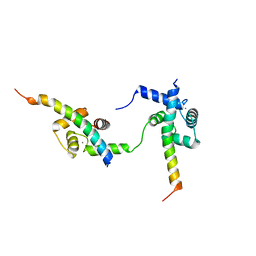 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
7RPP
 
 | | Crystal structure of human CEACAM1 with GFCC' and ABED face | | Descriptor: | 1,2-ETHANEDIOL, Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2021-08-04 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of human CEACAM1 oligomerization.
Commun Biol, 5, 2022
|
|
6O9T
 
 | | KirBac3.1 mutant at a resolution of 4.1 Angstroms | | Descriptor: | 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Inward rectifier potassium channel Kirbac3.1, POTASSIUM ION | | Authors: | Gulbis, J.M, Black, K.A, Miller, D.M. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
6APW
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 4-[(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)methyl]benzoic acid, Bifunctional ligase/repressor BirA | | Authors: | Cini, D.A, Wilce, M.C.J. | | Deposit date: | 2017-08-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
3FHP
 
 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-20 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
9GQO
 
 | | Structure of a Ca2+ bound phosphoenzyme intermediate in the inward-to-outward transition of Ca2+-ATPase 1 from Listeria monocytogenes | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase lmo0841, MAGNESIUM ION | | Authors: | Hansen, S.B, Flygaard, R.F, Kjaergaard, M, Nissen, P. | | Deposit date: | 2024-09-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the [Ca]E2P intermediate of Ca 2+ -ATPase 1 from Listeria monocytogenes.
Embo Rep., 26, 2025
|
|
8BHT
 
 | | ABCG2 turnover-1 state with tariquidar bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL, ... | | Authors: | Yu, Q, Kowal, J, Tajkhorshid, E, Locher, K.P. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential dynamics and direct interaction of bound ligands with lipids in multidrug transporter ABCG2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4RLU
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with 2',4,4'-trihydroxychalcone | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 2',4,4'-TRIHYDROXYCHALCONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
8B91
 
 | | Crystal structure of mutant PPARG (C313A) and NCOR2 with an inverse agonist (compound SI-1) | | Descriptor: | Nuclear receptor corepressor 2, Peroxisome proliferator-activated receptor gamma, ~{N}3-[4-[bis(fluoranyl)methoxy]-3-fluoranyl-phenyl]-4-chloranyl-6-fluoranyl-~{N}1-[(2-methoxyphenyl)methyl]benzene-1,3-dicarboxamide | | Authors: | Friberg, A, Orsi, D.L, Pook, E, Siegel, S, Lemke, C.T, Stellfeld, T, Puetter, V, Goldstein, J. | | Deposit date: | 2022-10-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of orally bioavailable 4-chloro-6-fluoroisophthalamides as covalent PPARG inverse-agonists.
Bioorg.Med.Chem., 78, 2022
|
|
