6W56
 
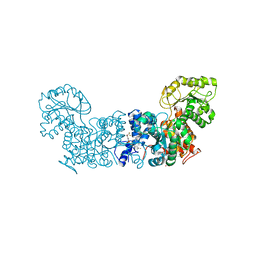 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC062) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme, ~{N}-[3-[[4-[bis(fluoranyl)methoxy]phenyl]sulfamoyl]phenyl]-3,5-bis(fluoranyl)benzamide | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
6OL8
 
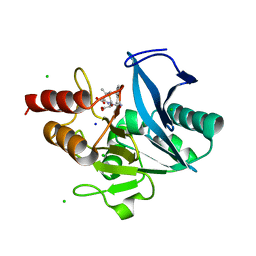 | | Crystal structure of NDM-12 metallo-beta-lactamase in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Metallo-beta-lactamase NDM-12, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
1H2W
 
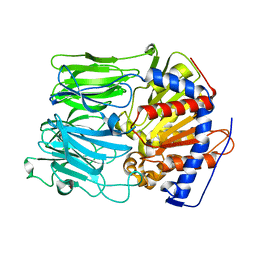 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN | | Descriptor: | GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Fulop, V. | | Deposit date: | 2002-08-20 | | Release date: | 2002-11-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Electrostatic Effects and Binding Determinants in the Catalysis of Prolyl Oligopeptidase: Site Specific Mutagenesis at the Oxyanion Binding Site
J.Biol.Chem., 277, 2002
|
|
1HH6
 
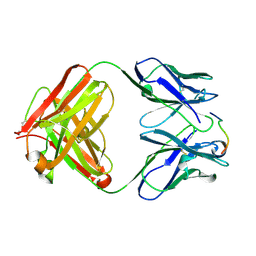 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
6WE8
 
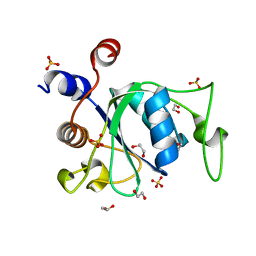 | | YTH domain of human YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-01 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Biochemical and structural basis for YTH domain of human YTHDC1 binding to methylated adenine in DNA.
Nucleic Acids Res., 48, 2020
|
|
6OBF
 
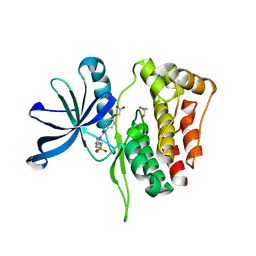 | | JAK2 JH2 in complex with JAK179 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6WTQ
 
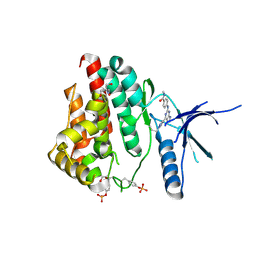 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-methyl-4-{[4-(1-propyl-1H-pyrazol-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}benzamide, ... | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79968476 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTN
 
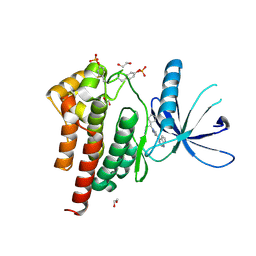 | | Human JAK2 JH1 domain in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, 1,2-ETHANEDIOL, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTP
 
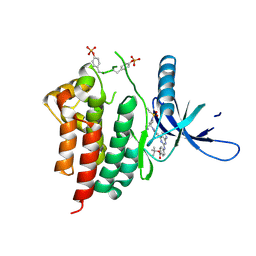 | | Human JAK2 JH1 domain in complex with PROTAC-intermediate linker handle 3 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, tert-butyl 4-[(4-{1-[3-(cyanomethyl)-1-(ethylsulfonyl)azetidin-3-yl]-1H-pyrazol-4-yl}-7H-pyrrolo[2,3-d]pyrimidin-2-yl)amino]benzoate | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6WTO
 
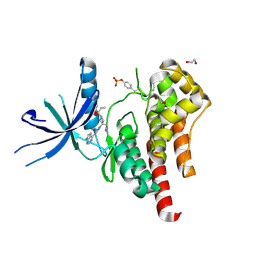 | | Human JAK2 JH1 domain in complex with Baricitinib | | Descriptor: | 1,2-ETHANEDIOL, Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Yu, S, Nithianantham, S, Fischer, M. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Degradation of Janus kinases in CRLF2-rearranged acute lymphoblastic leukemia.
Blood, 138, 2021
|
|
6X3A
 
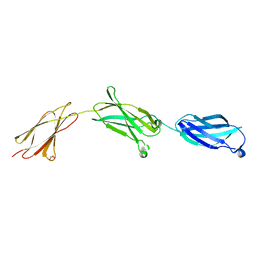 | |
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
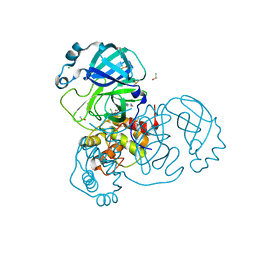 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6X39
 
 | |
6X38
 
 | |
7ANS
 
 | | Structure of SARS-CoV-2 Main Protease bound to Adrafinil. | | Descriptor: | 2-[(diphenylmethyl)-oxidanyl-$l^{3}-sulfanyl]-~{N}-oxidanyl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6WXL
 
 | |
6OCC
 
 | | JAK2 JH2 in complex with JAK190 | | Descriptor: | 2-[4-({5-amino-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenyl]-1,3-oxazole-4-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
7AVD
 
 | | Structure of SARS-CoV-2 Main Protease bound to SEN1269 ligand | | Descriptor: | 3-[[5-[3-(dimethylamino)phenoxy]pyrimidin-2-yl]amino]phenol, 3C-like proteinase, CHLORIDE ION | | Authors: | Koua, F, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Ewert, W, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AWU
 
 | | Structure of SARS-CoV-2 Main Protease bound to LSN2463359. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, ~{N}-propan-2-yl-5-(2-pyridin-4-ylethynyl)pyridine-2-carboxamide | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6OFD
 
 | | The crystal structure of octadecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P, Kan, M.W. | | Deposit date: | 2019-03-29 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
6OGO
 
 | | Crystal structure of NDM-9 metallo-beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raczynska, J.E, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2019-04-03 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Flexible loops of New Delhi metallo-beta-lactamase modulate its activity towards different substrates.
Int.J.Biol.Macromol., 158, 2020
|
|
7AQJ
 
 | | Structure of SARS-CoV-2 Main Protease bound to Triglycidyl isocyanurate. | | Descriptor: | 1-[(2~{R})-2-oxidanylpropyl]-3-[[(2~{R})-oxiran-2-yl]methyl]-5-[[(2~{S})-oxiran-2-yl]methyl]-1,3,5-triazinane-2,4,6-trione, 3C-like proteinase, Triglycidyl isocyanurate | | Authors: | Ewert, W, Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AOL
 
 | | Structure of SARS-CoV-2 Main Protease bound to Climbazole | | Descriptor: | (1~{S})-1-(4-chloranylphenoxy)-1-imidazol-1-yl-3,3-dimethyl-butan-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6XKL
 
 | | SARS-CoV-2 HexaPro S One RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrapp, D, Hsieh, C.-L, Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
Science, 369, 2020
|
|
