7W5Z
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial complex IV, composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-oxoglutarate/malate carrier protein, CARDIOLIPIN, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Letts, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-06 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7W7F
 
 | | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Li, X. | | Deposit date: | 2021-12-04 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis for modulation of human Na V 1.3 by clinical drug and selective antagonist.
Nat Commun, 13, 2022
|
|
7TJ8
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in nanodiscs | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7TJ9
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in GDN | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7SIP
 
 | | Structure of shaker-IR | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
7SJ1
 
 | | Structure of shaker-W434F | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | Deposit date: | 2021-10-15 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
7T3T
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3Q
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active B state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3U
 
 | | IP3, ATP, and Ca2+ bound type 3 IP3 receptor in the inactive state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3P
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active A state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7T3R
 
 | | IP3 and ATP bound type 3 IP3 receptor in the pre-active C state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Schmitz, E.A, Takahashi, H, Karakas, E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for activation and gating of IP 3 receptors.
Nat Commun, 13, 2022
|
|
7N16
 
 | | Structure of TAX-4_R421W apo closed state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, SODIUM ION | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N15
 
 | | Structure of TAX-4_R421W w/cGMP open state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7N17
 
 | | Structure of TAX-4_R421W apo open state | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel | | Authors: | Zheng, X, Li, H, Hu, Z, Su, D, Yang, J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characterization of an achromatopsia-associated mutation in a phototransduction channel.
Commun Biol, 5, 2022
|
|
7TDG
 
 | |
7TDI
 
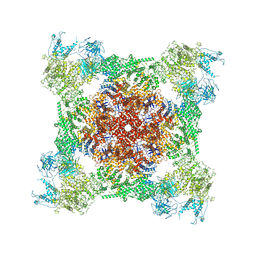 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 2 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDH
 
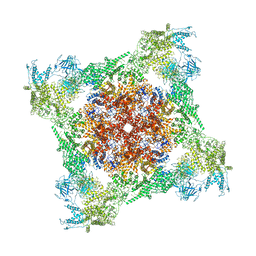 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in open conformation | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDK
 
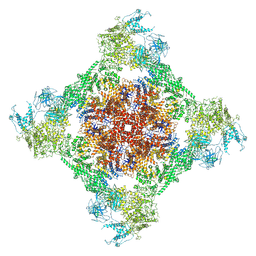 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 3 (Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDJ
 
 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 1(Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7PHI
 
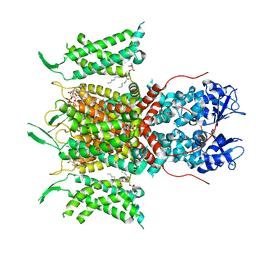 | | Human voltage-gated potassium channel Kv3.1 (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Marsden, B, MacLean, E.M, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHK
 
 | | Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHH
 
 | | Human voltage-gated potassium channel Kv3.1 (apo condition) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Marsden, B, MacLean, E.M, Fernandez-Cid, A, Pike, A.C.W, Savva, C, Ragan, T.J, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7PHL
 
 | | Human voltage-gated potassium channel Kv3.1 (with EDTA) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, POTASSIUM ION, Potassium voltage-gated channel, ... | | Authors: | Chi, G, Qian, P, Castro-Hartmann, P, Venkaya, S, Singh, N.K, McKinley, G, Mukhopadhyay, S.M.M, Fernandez-Cid, A, Pike, A.C.W, Marsden, B, MacLean, E.M, Sader, K, Burgess-Brown, N.A, Duerr, K.L. | | Deposit date: | 2021-08-17 | | Release date: | 2022-03-02 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human Kv3.1 channel reveals gating control by the cytoplasmic T1 domain.
Nat Commun, 13, 2022
|
|
7RHS
 
 | | Cryo-EM structure of apo-state of human CNGA3/CNGB3 channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cyclic nucleotide-gated cation channel alpha-3, Cyclic nucleotide-gated cation channel beta-3, ... | | Authors: | Zheng, X, Yang, J. | | Deposit date: | 2021-07-18 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7WF3
 
 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
