9CMH
 
 | |
1NCC
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IGG2A-KAPPA NC41 FAB (HEAVY CHAIN), ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
4JDD
 
 | |
5UK0
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UK1
 
 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
4JC3
 
 | |
1NCB
 
 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
1RAL
 
 | | THREE-DIMENSIONAL STRUCTURE OF RAT LIVER 3ALPHA-HYDROXYSTEROID(SLASH)DIHYDRODIOL DEHYDROGENASE: A MEMBER OF THE ALDO-KETO REDUCTASE SUPERFAMILY | | Descriptor: | 3-ALPHA-HYDROXYSTEROID DEHYDROGENASE | | Authors: | Hoog, S.S, Pawlowski, J.E, Alzari, P.M, Penning, T.M, Lewis, M. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase: a member of the aldo-keto reductase superfamily.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
5UK2
 
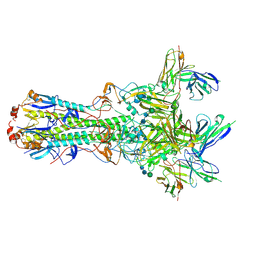 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
5UJZ
 
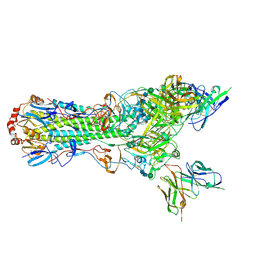 | | CryoEM structure of an influenza virus receptor-binding site antibody-antigen interface - Class 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Liu, Y, Pan, J, Caradonna, T, Jenni, S, Raymond, D.D, Schmidt, A.G, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2017-01-19 | | Release date: | 2017-05-31 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structure of an Influenza Virus Receptor-Binding Site Antibody-Antigen Interface.
J. Mol. Biol., 429, 2017
|
|
7OGS
 
 | |
4EN0
 
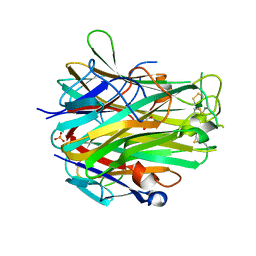 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
5G08
 
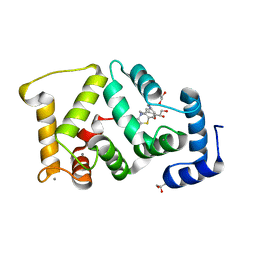 | | Crystal structure of Drosophila NCS-1 bound to chlorpromazine | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7OOT
 
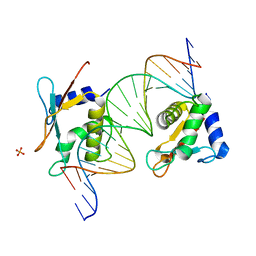 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element | | Descriptor: | DNA (5'-D(P*AP*GP*CP*TP*TP*TP*CP*TP*CP*GP*GP*TP*TP*TP*CP*AP*GP*TP*TP*G)-3'), DNA (5'-D(P*TP*CP*AP*AP*CP*TP*GP*AP*AP*AP*CP*CP*GP*AP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor 4, ... | | Authors: | Agnarelli, A, El Omari, K, Alt, A.O, Mancini, E.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to interferon-stimulated response element
To Be Published
|
|
4BED
 
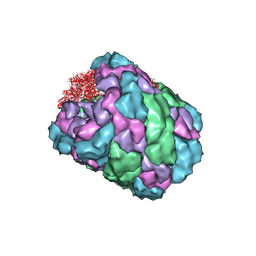 | |
5FYX
 
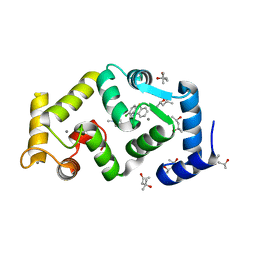 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD16 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, FREQUENIN 2, ... | | Authors: | Martinez-Gonzalez, L, Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2016-03-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5AAN
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4KG8
 
 | | Crystal structure of light mutant | | Descriptor: | Tumor necrosis factor ligand superfamily member 14 | | Authors: | Liu, W, Zhan, C, Kumar, P.R, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-28 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
1JTT
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, Lysozyme, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JTO
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | Lysozyme, Vh Single-Domain Antibody | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
7AP7
 
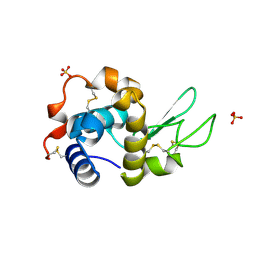 | | Structure of the W64R amyloidogenic variant of human lysozyme | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Vettore, N, Herman, R, Kerff, F, Charlier, P, Sauvage, E, Brans, A, Morray, J, Dobson, C, Kumita, J, Dumoulin, M. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of the structural, dynamic and aggregation properties of the W64R amyloidogenic variant of human lysozyme.
Biophys.Chem., 271, 2021
|
|
4KGG
 
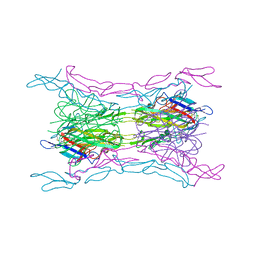 | | Crystal structure of light mutant2 and dcr3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Bonanno, J.B, Zhan, C, Kumar, P.R, Toro, R, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4J6G
 
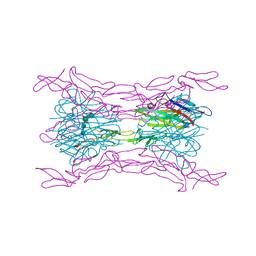 | | CRYSTAL STRUCTURE OF LIGHT AND DcR3 COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-11 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
5LZT
 
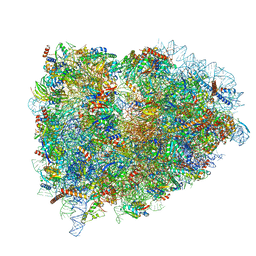 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
