2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2D1B
 
 | | Solution RNA structure model of the HIV-1 dimerization initiation site in the kissing-loop dimer | | Descriptor: | RNA | | Authors: | Baba, S, Takahashi, K, Noguchi, S, Takaku, H, Koyanagi, Y, Yamamoto, N, Kawai, G. | | Deposit date: | 2005-08-15 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.
J.Biochem.(Tokyo), 138, 2005
|
|
2DDR
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with calcium ion | | Descriptor: | CALCIUM ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
2CYA
 
 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
3VUB
 
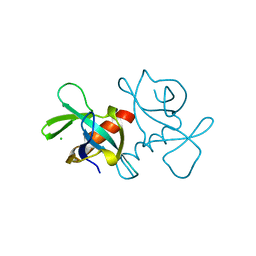 | | CCDB, A TOPOISOMERASE POISON FROM E. COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
7CPN
 
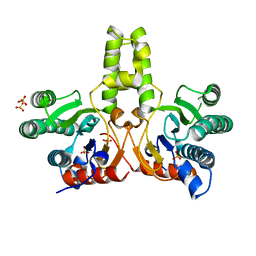 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | GLYCEROL, SULFATE ION, Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahasi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7DDT
 
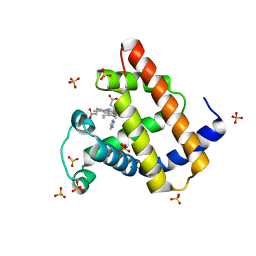 | | Ancestral myoglobin aMbSe of Enaliarctos relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSe, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDR
 
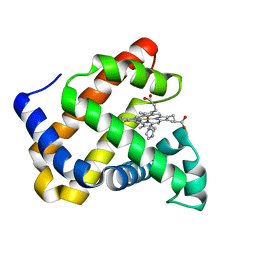 | | Ancestral myoglobin aMbSp of Puijila Darwini relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDS
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative | | Descriptor: | Ancestral myoglobin aMbSp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDU
 
 | | Elephant seal myoglobin esMb | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
1F54
 
 | | SOLUTION STRUCTURE OF THE APO N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
1F55
 
 | | SOLUTION STRUCTURE OF THE CALCIUM BOUND N-TERMINAL DOMAIN OF YEAST CALMODULIN | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Ishida, H, Takahashi, K, Nakashima, K, Kumaki, Y, Nakata, M, Hikichi, K, Yazawa, M. | | Deposit date: | 2000-06-13 | | Release date: | 2003-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the N-terminal Domain of Yeast Calmodulin:
Ca2+-Dependent Conformational Change and Its Functional Implication
Biochemistry, 39, 2000
|
|
7XAR
 
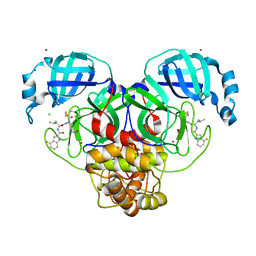 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
7Y8P
 
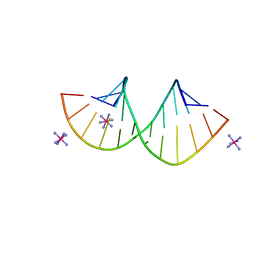 | | Crystal structure of 4'-selenoRNA duplex | | Descriptor: | COBALT HEXAMMINE(III), RNA (5'-R(*GP*GP*AP*(IKS)P*(ILK)P*(IKS)P*GP*AP*GP*UP*CP*C)-3') | | Authors: | Kondo, J, Minakawa, N, Ohta, M, Takahashi, H, Tarashima, N. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and properties of fully-modified 4'-selenoRNA, an endonuclease-resistant RNA analog.
Bioorg.Med.Chem., 76, 2022
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
6KBI
 
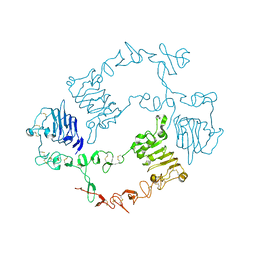 | | Crystal structure of ErbB3 N418Q mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-3 | | Authors: | Kato, K, Yao, M. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of ErbB3 N418Q mutant
To Be Published
|
|
7ZCJ
 
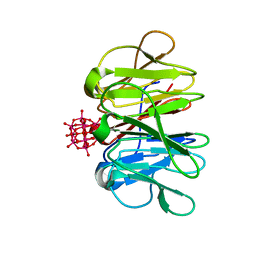 | | Crystal structure of Pizza6-TNH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
8E35
 
 | | E. coli 50S ribosome bound to compound SAB002 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E33
 
 | | E. coli 50S ribosome bound to compound streptogramin analog SAB001 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L15, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
8E32
 
 | | E. coli 50S ribosome bound to compound streptogramin analogs SA1 and SB1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-yn-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Potential for clicked streptogramin analogs
To Be Published
|
|
1IPJ
 
 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS COMPLEXES WITH N-ACETYL-D-GLUCOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-CONGLYCININ, BETA CHAIN | | Authors: | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | Deposit date: | 2001-05-16 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
7ZPZ
 
 | | Crystal structure of Pizza6-TSR-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TSR-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
7ZQ2
 
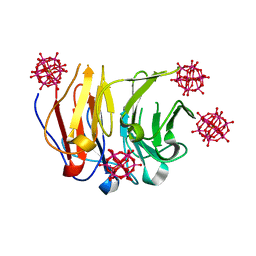 | | Crystal structure of Pizza6-RSH-TSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-RSH-TSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
