1MPG
 
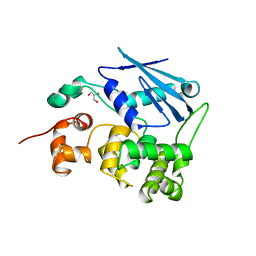 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
1MWJ
 
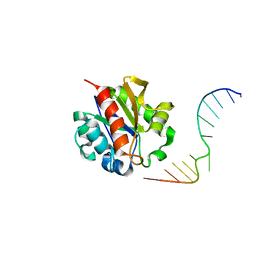 | | Crystal Structure of a MUG-DNA pseudo substrate complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*A*GP*(DU)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Scharer, O, Savva, R, Brown, T, Jiricny, J, Verdine, G.L, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of a thwarted mismatch glycosylase DNA repair complex
Embo J., 18, 1999
|
|
6D92
 
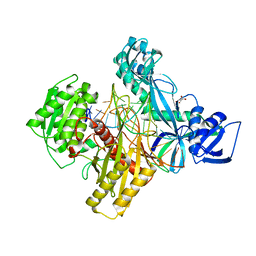 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-A non-canonical pair at position 3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*TP*GP*CP*AP*GP*AP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-27 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
4ZCF
 
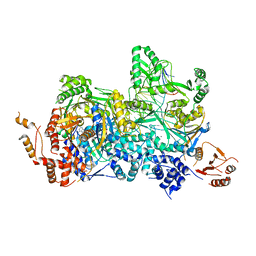 | | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, DNA 20-mer AATCATAGTCTACTGCTGTA, ... | | Authors: | Gupta, Y.K, Chan, S.H, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I.
Nat Commun, 6, 2015
|
|
6GDS
 
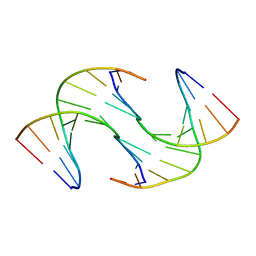 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
7N3P
 
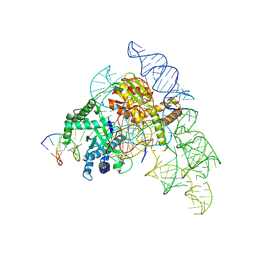 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | Descriptor: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
8YFJ
 
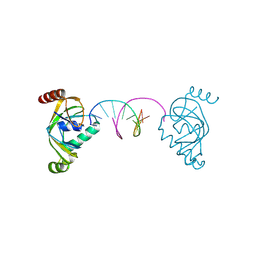 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8B6E
 
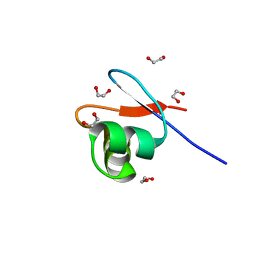 | | crystal structure of the DNA-binding short chromatophore-targeted protein sCTP-23166 from Paulinella chromatophora | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, sCTP-23166 | | Authors: | Macorano, L, Applegate, V, Hoeppner, A, Smits, S.H.J, Nowack, E.C.M. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | DNA-binding and protein structure of nuclear factors likely acting in genetic information processing in the Paulinella chromatophore.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8YFI
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
3BI3
 
 | |
4QC9
 
 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
6FAS
 
 | | Crystal structure of VAL1 B3 domain in complex with cognate DNA | | Descriptor: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*GP*CP*CP*AP*TP*GP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*CP*GP*GP*TP*GP*CP*AP*TP*GP*GP*CP*T)-3') | | Authors: | Sasnauskas, G. | | Deposit date: | 2017-12-17 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of DNA target recognition by the B3 domain of Arabidopsis epigenome reader VAL1.
Nucleic Acids Res., 46, 2018
|
|
7KQM
 
 | | Binary complex of TERT (telomerase reverse transcriptase) with RNA/telomeric DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*GP*CP*AP*CP*CP*TP*G)-3'), ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1LEB
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
6F1K
 
 | | Structure of ARTD2/PARP2 WGR domain bound to double strand DNA without 5'phosphate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*GP*CP*CP*TP*AP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*TP*AP*GP*GP*C)-3'), GLYCEROL, ... | | Authors: | Obaji, E, Haikarainen, T, Lehtio, L. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for DNA break recognition by ARTD2/PARP2.
Nucleic Acids Res., 46, 2018
|
|
3BSE
 
 | | Crystal structure analysis of a 16-base-pair B-DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DA*DCP*DAP*DCP*DTP*DAP*DCP*DAP*DAP*DTP*DGP*DTP*DTP*DGP*DCP*DAP*DAP*DT)-3'), DNA (5'-D(*DG*DTP*DAP*DTP*DTP*DGP*DCP*DAP*DAP*DCP*DAP*DTP*DTP*DGP*DTP*DAP*DGP*DT)-3') | | Authors: | Narayana, N. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Analysis of a Sex-Specific Enhancer Element: Sequence-Dependent DNA Structure, Hydration, and Dynamics
J.Mol.Biol., 385, 2009
|
|
3BKZ
 
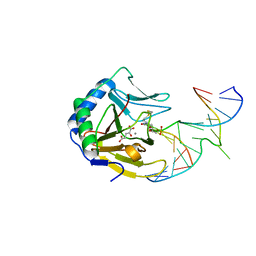 | | X-ray structure of E coli AlkB crosslinked to dsDNA in the active site | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB, DNA (5'-D(*DAP*DAP*DCP*DGP*DAP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DT)-3'), ... | | Authors: | Yi, C, Yang, C.-G, He, C. | | Deposit date: | 2007-12-09 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA
Nature, 452, 2008
|
|
102D
 
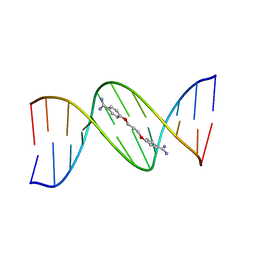 | |
3BTX
 
 | | X-ray structure of human ABH2 bound to dsDNA through active site cross-linking | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), DNA (5'-D(*DTP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DAP*DTP*DAP*DCP*DA)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
3BU0
 
 | | crystal structure of human ABH2 cross-linked to dsDNA with cofactors | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA (5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Yang, C.-G, Yi, C, He, C. | | Deposit date: | 2007-12-31 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA.
Nature, 452, 2008
|
|
6IFM
 
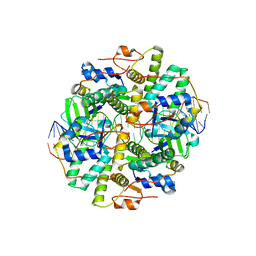 | | Crystal structure of DNA bound VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, DNA backward (27-MER), DNA forward (27-MER), ... | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
5NFV
 
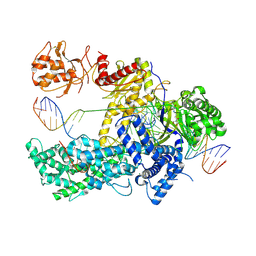 | | Crystal structure of catalytically inactive FnCas12 mutant bound to an R-loop structure containing a pre-crRNA mimic and full-length DNA target | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CRISPR-associated endonuclease Cpf1, DNA non-target strand, ... | | Authors: | Swarts, D.C, van der Oost, J, Jinek, M. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a.
Mol. Cell, 66, 2017
|
|
1MW8
 
 | |
7MG5
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-ATAT | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
7MG9
 
 | | Concanavalin A bound to DNA glycoconjugates, Man-TTTT and Man-AAAA | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
