3D80
 
 | | Structural Analysis of a Holo Enzyme Complex of Mouse Dihydrofolate Reductase with NADPH and a Ternary Complex wtih the Potent and Selective Inhibitor 2,4-Diamino-6-(2'-hydroxydibenz[b,f]azepin-5-yl)methylpteridine | | Descriptor: | (4aS)-5-[(2,4-diaminopteridin-6-yl)methyl]-4a,5-dihydro-2H-dibenzo[b,f]azepin-8-ol, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Cody, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of a holoenzyme complex of mouse dihydrofolate reductase with NADPH and a ternary complex with the potent and selective inhibitor 2,4-diamino-6-(2'-hydroxydibenz[b,f]azepin-5-yl)methylpteridine.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3D1Q
 
 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 400 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
3RW6
 
 | | Structure of nuclear RNA export factor TAP bound to CTE RNA | | Descriptor: | Nuclear RNA export factor 1, constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | | Authors: | Teplova, M, Khin, N.W, Patel, D.J, Izaurralde, E. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3D66
 
 | |
3RVO
 
 | | Structure of CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Y | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
2RVA
 
 | |
3CHN
 
 | | Solution structure of human secretory IgA1 | | Descriptor: | Ig alpha-1 chain C region, Immunoglobulin kappa light chain, Secretory component | | Authors: | Bonner, A, Almogren, A, Furtado, P.B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2008-03-10 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity.
Mucosal Immunol, 2, 2009
|
|
3RVM
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D and E89R | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3COL
 
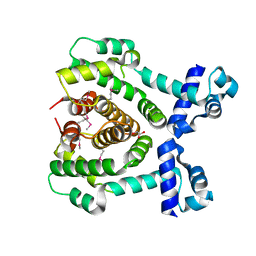 | |
3RUJ
 
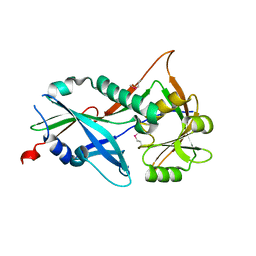 | |
3CQ3
 
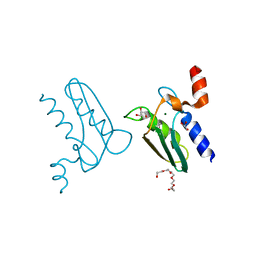 | | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8 | | Descriptor: | GLYCEROL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Satoh, S, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-02 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the DTDP-4-Keto-L-Rhamnose Reductase related protein (other form) from Thermus Thermophilus HB8
To be Published
|
|
2RV9
 
 | |
3CUQ
 
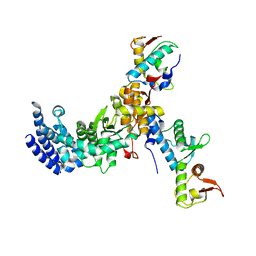 | |
3CI0
 
 | |
3RYB
 
 | | Lactococcal OppA complexed with SLSQSLSQS | | Descriptor: | Oligopeptide, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Importance of a Hydrophobic Pocket for Peptide Binding in Lactococcal OppA.
J.Bacteriol., 193, 2011
|
|
2RIN
 
 | | ABC-transporter choline binding protein in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
3CK5
 
 | |
3CZX
 
 | |
3RZE
 
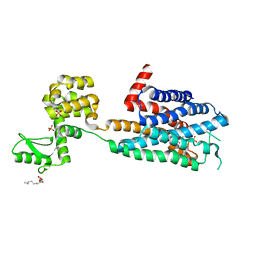 | | Structure of the human histamine H1 receptor in complex with doxepin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (3E)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, (3Z)-3-(dibenzo[b,e]oxepin-11(6H)-ylidene)-N,N-dimethylpropan-1-amine, ... | | Authors: | Shimamura, T, Han, G.W, Shiroishi, M, Weyand, S, Tsujimoto, H, Winter, G, Katritch, V, Abagyan, R, Cherezov, V, Liu, W, Kobayashi, T, Stevens, R, Iwata, S, GPCR Network (GPCR) | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the human histamine H1 receptor complex with doxepin.
Nature, 475, 2011
|
|
2RHF
 
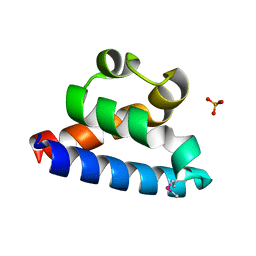 | | D. radiodurans RecQ HRDC domain 3 | | Descriptor: | DNA helicase RecQ, PHOSPHATE ION | | Authors: | Keck, J.L, Killoran, M.P. | | Deposit date: | 2007-10-09 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ.
Nucleic Acids Res., 36, 2008
|
|
2RHZ
 
 | |
3D67
 
 | |
2ROP
 
 | | Solution structure of domains 3 and 4 of human ATP7B | | Descriptor: | Copper-transporting ATPase 2 | | Authors: | Banci, L, Bertini, I, Cantini, F, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1
Biochemistry, 47, 2008
|
|
3FN1
 
 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
3F87
 
 | |
