9MFQ
 
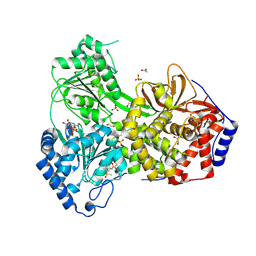 | | Cat DHX9 in Complex with Compound 10 and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lockbaum, G.J, Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A, Grigoriu, S. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of ATX968: An Orally Available Allosteric Inhibitor of DHX9.
J.Med.Chem., 68, 2025
|
|
7OKB
 
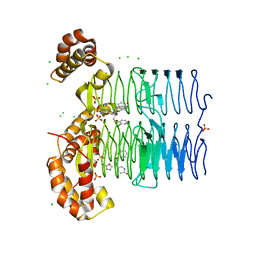 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 45 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CHLORIDE ION, SULFATE ION, ... | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OEK
 
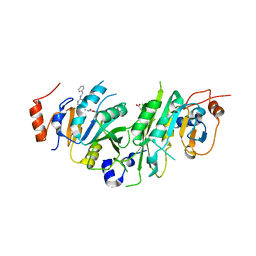 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ091 | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-2-oxidanyl-N-[[(3R)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7OK2
 
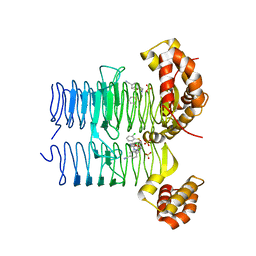 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
7OEJ
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ090 | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3R)-1-[6-(methylamino)pyrimidin-4-yl]-3-oxidanyl-piperidin-3-yl]methyl]-2-oxidanyl-benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
5A0D
 
 | | N-terminal thioester domain of surface protein from Clostridium perfringens, Cys138Ala mutant | | Descriptor: | 1,2-ETHANEDIOL, SURFACE ANCHORED PROTEIN | | Authors: | Walden, M, Edwards, J.M, Dziewulska, A.M, Kan, S.-Y, Schwarz-Linek, U, Banfield, M.J. | | Deposit date: | 2015-04-17 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An internal thioester in a pathogen surface protein mediates covalent host binding.
Elife, 4, 2015
|
|
7OEH
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ059b | | Descriptor: | 4-[[(3S)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3S)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-05-03 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7A58
 
 | |
6ONW
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6X0O
 
 | | Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB | | Authors: | Tan, Y.Z, Rodrigues, J, Keener, J.E, Zheng, R.B, Brunton, R, Kloss, B, Giacometti, S.I, Rosario, A.L, Zhang, L, Niederweis, M, Clarke, O.B, Lowary, T.L, Marty, M.T, Archer, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Nat Commun, 11, 2020
|
|
5FAA
 
 | | Crystal structure of C-terminal domain of shaft pilin spaA from Lactobacillus rhamnosus GG, - I422 space group | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
302D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL IN' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | Descriptor: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | Deposit date: | 1996-06-26 | | Release date: | 1997-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
8X9Q
 
 | | Cte-A695U-branching,Mg,active | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (809-MER) | | Authors: | Ling, X.B, Ma, J.B. | | Deposit date: | 2023-11-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of a natural circularly permuted group II intron reveal mechanisms of branching and backsplicing.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6ZV9
 
 | | Terbium(III)-bound de novo TIM barrel-ferredoxin fold fusion dimer with 4-glutamate binding site and tryptophan antenna (TFD-EE N6W) | | Descriptor: | 1,2-ETHANEDIOL, TERBIUM(III) ION, TFD-EE | | Authors: | Caldwell, S, Haydon, I, Piperidou, N, Huang, P, Hilvert, D, Baker, D, Zeymer, C. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PTI
 
 | |
5V2G
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, 20-mer Peptide | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-03 | | Release date: | 2017-09-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VS7
 
 | | Crystal structure of ADI-19425 Fab in complex with anti-idiotypic 2C1 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2C1 Heavy Chain, 2C1 Light Chain, ... | | Authors: | Kher, G, Homad, L.J, McGuire, A.T, Pancera, M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A novel RSV vaccine candidate derived from anti-idiotypic antibodies targets putative neutralizing B-cells in bulk PBMCs
To Be Published
|
|
8EYU
 
 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8Y54
 
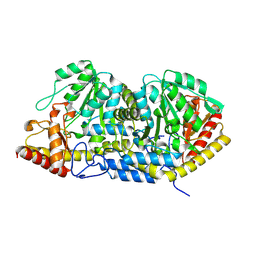 | |
7Y2P
 
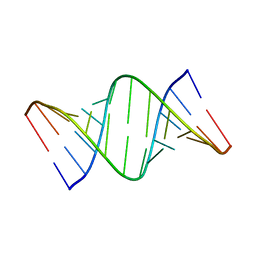 | |
1DCG
 
 | | THE MOLECULAR STRUCTURE OF THE LEFT-HANDED Z-DNA DOUBLE HELIX AT 1.0 ANGSTROM ATOMIC RESOLUTION. GEOMETRY, CONFORMATION, AND IONIC INTERACTIONS OF D(CGCGCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Gessner, R.V, Frederick, C.A, Quigley, G.J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The molecular structure of the left-handed Z-DNA double helix at 1.0-A atomic resolution. Geometry, conformation, and ionic interactions of d(CGCGCG).
J.Biol.Chem., 264, 1989
|
|
483D
 
 | |
444D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
442D
 
 | | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', BENZIMIDAZOLE DERIVATIVE COMPLEX | | Descriptor: | 2'-(3-IODO-4-METHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Clark, G.R, Squire, C.J, Martin, R.F, White, J. | | Deposit date: | 1999-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of m-iodo Hoechst-DNA complexes in crystals with reduced solvent content: implications for minor groove binder drug design.
Nucleic Acids Res., 28, 2000
|
|
6MSA
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 7,8-dimethoxy-1-methyl-2H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
