7K5Y
 
 | |
7K60
 
 | |
7C7X
 
 | |
7BP4
 
 | |
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
6M2M
 
 | |
7CRP
 
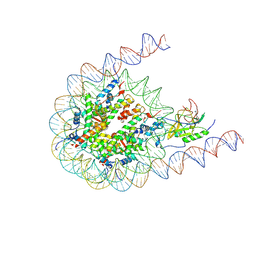 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
6USJ
 
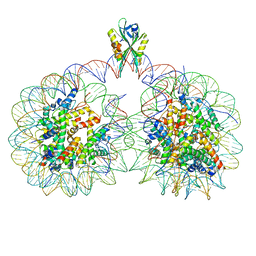 | | Structure of two nucleosomes bridged by human PARP2 | | Descriptor: | Histone H2A, Histone H2B type 1-J, Histone H3.1, ... | | Authors: | Gaullier, G, Bowerman, S, Luger, K. | | Deposit date: | 2019-10-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Bridging of nucleosome-proximal DNA double-strand breaks by PARP2 enhances its interaction with HPF1.
Plos One, 15, 2020
|
|
7CCR
 
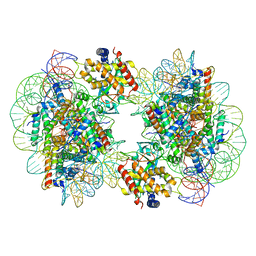 | | Structure of the 2:2 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
7CCQ
 
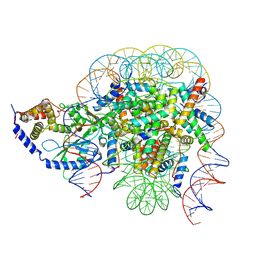 | | Structure of the 1:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Cao, D, Han, X, Fan, X, Xu, R.M, Zhang, X. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for nucleosome-mediated inhibition of cGAS activity.
Cell Res., 30, 2020
|
|
6LA8
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6LA9
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6M44
 
 | | 355 bp di-nucleosome harboring cohesive DNA termini (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6M3V
 
 | | 355 bp di-nucleosome harboring cohesive DNA termini | | Descriptor: | CALCIUM ION, DNA (355-MER), Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2020-03-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6R0N
 
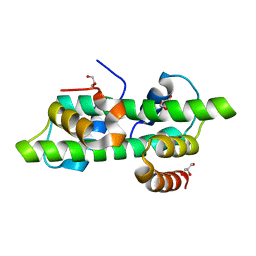 | | Histone fold domain of AtNF-YB2/NF-YC3 in I2 | | Descriptor: | GLYCEROL, NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Bernardini, A, Fornara, F, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R0L
 
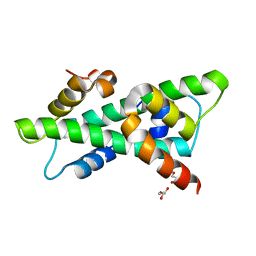 | |
6R0M
 
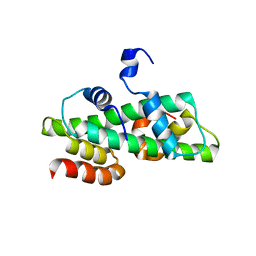 | | Histone fold domain of AtNF-YB2/NF-YC3 in P212121 | | Descriptor: | NF-YB2, NF-YC3 | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6R2V
 
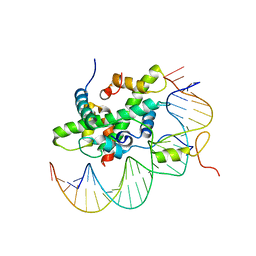 | | Arabidopsis NF-Y/CCAAT-box complex | | Descriptor: | FT (-5kb) CCAAT-box 3', FT (-5kb) CCAAT-box 5', NF-YB2, ... | | Authors: | Chaves-Sanjuan, A, Gnesutta, N, Chiara, M, Bernardini, A, Fornara, F, Horner, D, Nardini, M, Mantovani, R. | | Deposit date: | 2019-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural determinants for NF-Y subunit organization and NF-Y/DNA association in plants.
Plant J., 105, 2021
|
|
6Y5E
 
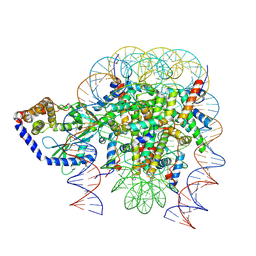 | | Structure of human cGAS (K394E) bound to the nucleosome (focused refinement of cGAS-NCP subcomplex) | | Descriptor: | Cyclic GMP-AMP synthase, DNA (153-MER), Histone H2A type 2-C, ... | | Authors: | Pathare, G.R, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural mechanism of cGAS inhibition by the nucleosome.
Nature, 587, 2020
|
|
6M4D
 
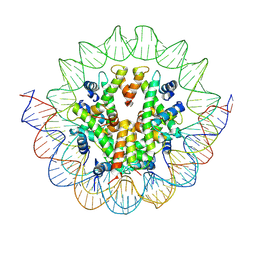 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (125-MER), Histone H2A.V, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
