5WWE
 
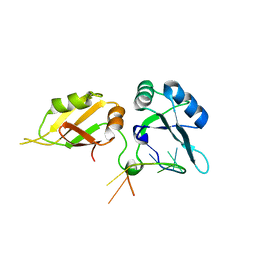 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*GP*AP*CP*UP*AP*GP*A)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
6EXN
 
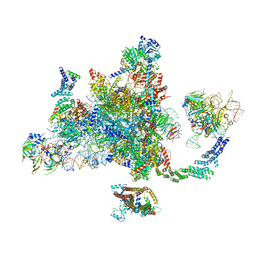 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
5WWG
 
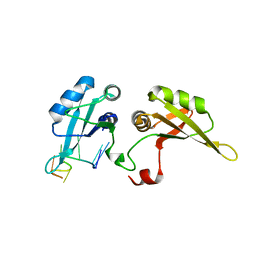 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWF
 
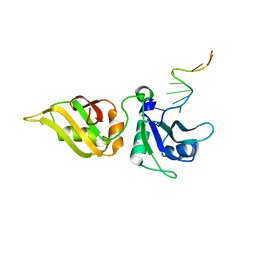 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
6EM1
 
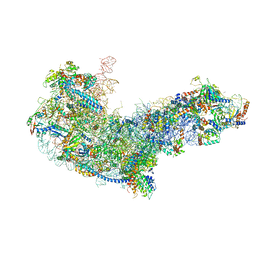 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM3
 
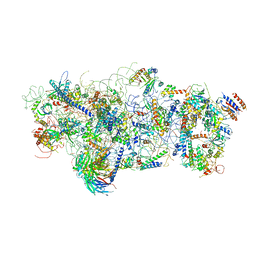 | | State A architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM4
 
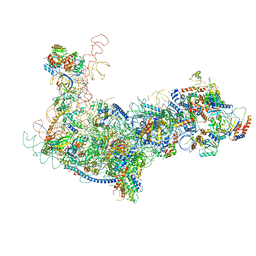 | | State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EM5
 
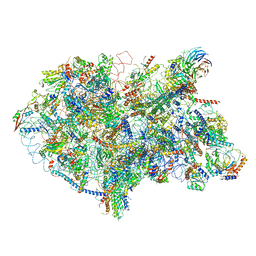 | | State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6ELZ
 
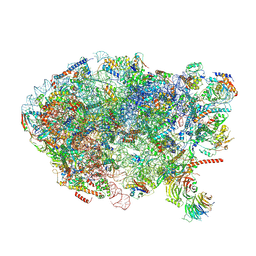 | | State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-09-30 | | Release date: | 2017-12-27 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
5X3Z
 
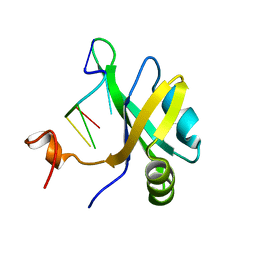 | | Solution structure of musashi1 RBD2 in complex with RNA | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5X3Y
 
 | | Refined solution structure of musashi1 RBD2 | | Descriptor: | RNA-binding protein Musashi homolog 1 | | Authors: | Iwaoka, R, Nagata, T, Tsuda, K, Imai, T, Okano, H, Kobayashi, N, Katahira, M. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Recognition of r(UAG) by Musashi-1 RBD2, and Construction of a Model of Musashi-1 RBD1-2 Bound to the Minimum Target RNA
Molecules, 22, 2017
|
|
5OPT
 
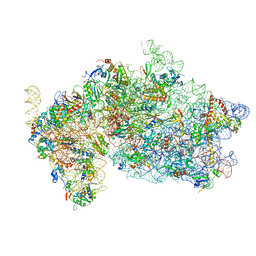 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5OSG
 
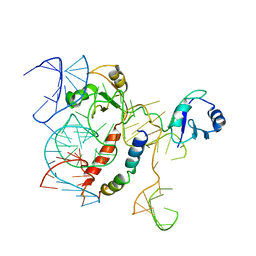 | | Structure of KSRP in context of Leishmania donovani 80S | | Descriptor: | 18S rRNA, 40S ribosomal protein S6, RNA binding protein, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5OOB
 
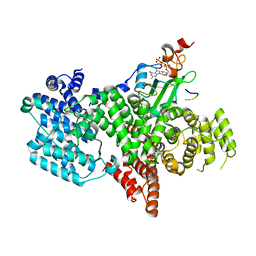 | |
5OO6
 
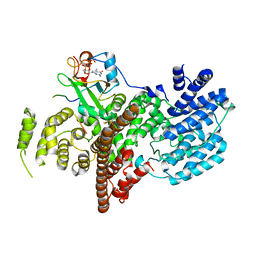 | | Complex of human nuclear cap-binding complex with ARS2 C-terminal peptide | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Cusack, S, Schulze, W.M. | | Deposit date: | 2017-08-06 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for mutually exclusive co-transcriptional nuclear cap-binding complexes with either NELF-E or ARS2.
Nat Commun, 8, 2017
|
|
5M8I
 
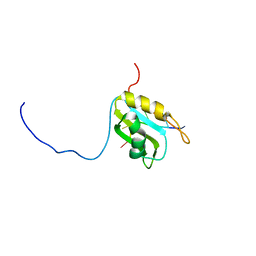 | |
5SZW
 
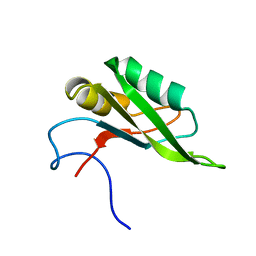 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
5KWQ
 
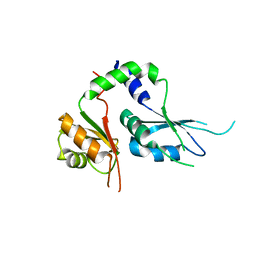 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KW6
 
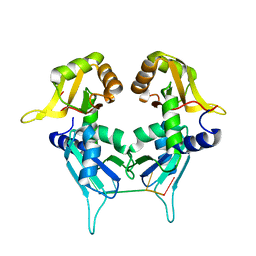 | | Two Tandem RRM Domains of PUF60 Bound to an AdML Pre-mRNA 3' Splice Site Analogue with a Modified Binding-Site Nucleic Acid Base | | Descriptor: | DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KW1
 
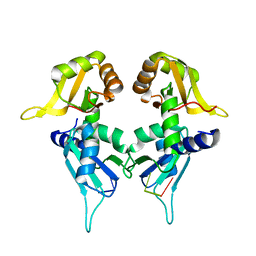 | | Crystal Structure of the Two Tandem RRM Domains of PUF60 Bound to a Modified AdML Pre-mRNA 3' Splice Site Analogue | | Descriptor: | CHLORIDE ION, DNA/RNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KVY
 
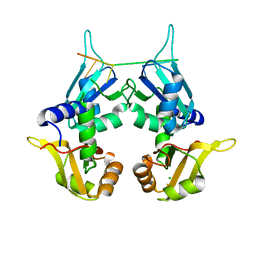 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | Descriptor: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5O9Z
 
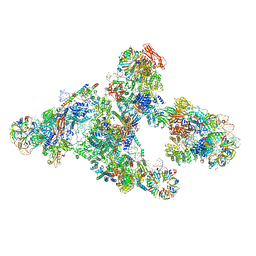 | |
5IFN
 
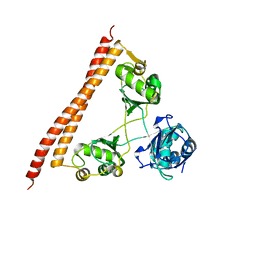 | |
5O1X
 
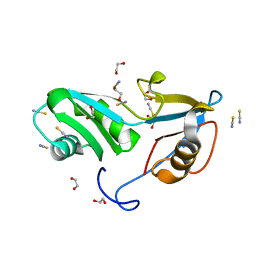 | | Structure of Nrd1 RNA binding domain | | Descriptor: | 1,2-ETHANEDIOL, Protein NRD1, THIOCYANATE ION | | Authors: | Franco-Echevarria, E, Perez-Canadillas, J.M, Gonzalez, B. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition.
Nucleic Acids Res., 45, 2017
|
|
5O1W
 
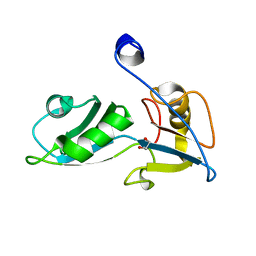 | |
