4AF1
 
 | | Archeal Release Factor aRF1 | | Descriptor: | PEPTIDE CHAIN RELEASE FACTOR SUBUNIT 1, ZINC ION | | Authors: | Karlsen, J.L, Kjeldgaard, M. | | Deposit date: | 2012-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Archeal Release Factor Arf1 Contains a Metal Binding Domain
To be Published
|
|
4AL6
 
 | |
3MD0
 
 | |
4NFZ
 
 | | Crystal structure of polymerase subunit PA N-terminal endonuclease domain from bat-derived influenza virus H17N10 | | Descriptor: | MANGANESE (II) ION, Polymerase PA | | Authors: | Tefsen, B, Lu, G, Zhu, Y, Haywood, J, Zhao, L, Deng, T, Qi, J, Gao, G.F. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N-Terminal Domain of PA from Bat-Derived Influenza-Like Virus H17N10 Has Endonuclease Activity
J.Virol., 88, 2014
|
|
3MM6
 
 | | Dissimilatory sulfite reductase cyanide complex | | Descriptor: | CYANIDE ION, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
4NTF
 
 | |
4HG0
 
 | | Crystal Structure of magnesium and cobalt efflux protein CorC, Northeast Structural Genomics Consortium (NESG) Target ER40 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein CorC | | Authors: | Kuzin, A, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Cooper, B, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Northeast Structural Genomics Consortium Target ER40
To be Published
|
|
4HGA
 
 | | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Liu, C.P, Xiong, C.Y, Wang, M.Z, Yu, Z.L, Yang, N, Chen, P, Zhang, Z.G, Li, G.H, Xu, R.M. | | Deposit date: | 2012-10-07 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4HHA
 
 | | Anti-Human Cytomegalovirus (HCMV) Fab KE5 with epitope peptide AD-2S1 | | Descriptor: | Antibody KE5, CHLORIDE ION, Fab KE5, ... | | Authors: | Bryson, S, Risnes, L, Damgupta, S, Thomson, C.A, Pfoh, R, Schrader, J.W, Pai, E.F. | | Deposit date: | 2012-10-09 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of Preferred Human IgV Genes-Based Protective Antibodies Identify How Conserved Residues Contact Diverse Antigens and Assign Source of Specificity to CDR3 Loop Variation.
J. Immunol., 196, 2016
|
|
3MPZ
 
 | |
4D5O
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E212Q soaked with xylopentaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
3MNW
 
 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment with a gp41 MPER-derived peptide in a helical conformation | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
3MOB
 
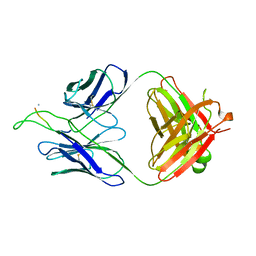 | | Crystal structure of the neutralizing HIV antibody 2F5 Fab fragment (recombinantly produced Fab) with 11 aa gp41 MPER-derived peptide | | Descriptor: | ANTI-HIV-1 ANTIBODY 2F5 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 2F5 LIGHT CHAIN, gp41 MPER-derived peptide | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a Non-Neutralizing HIV-1 gp41 Envelope Antibody Demonstrates Neutralization Mechanism of gp41 Antibodies
To be Published
|
|
4GTB
 
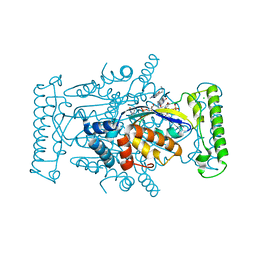 | | T. Maritima FDTS with FAD, dUMP, and Raltitrexed. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HFK
 
 | |
3MAS
 
 | |
4D2S
 
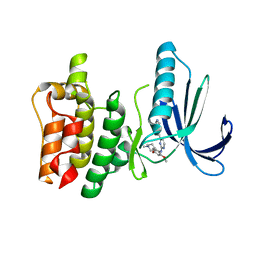 | | Human TTK in complex with a Dyrk1B inhibitor | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, N-{2-methoxy-4-[(1-methylpiperidin-4-yl)oxy]phenyl}-4-(1H-pyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine | | Authors: | Debreczeni, J.E, Kettle, J.G, Ballard, P, Bardelle, C, Butterworth, S, Colclough, N, Critchlow, S.E, Fairley, G, Fillery, S, Graham, M.A, Goodwin, L, Guichard, S, Hudson, K, Mahmood, A, Vincent, J, Ward, R.A, Whittaker, D. | | Deposit date: | 2014-05-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Optimization of a Novel Series of Dyrk1B Kinase Inhibitors to Explore a Mek Resistance Hypothesis.
J.Med.Chem., 58, 2015
|
|
3MAB
 
 | | CRYSTAL STRUCTURE OF AN UNCHARACTERIZED PROTEIN FROM LISTERIA MONOCYTOGENES, Triclinic FORM | | Descriptor: | uncharacterized protein | | Authors: | Madegowda, M, Chruszcz, M, Minor, W, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of an uncharacterized protein from listeria monocytogenes
To be Published
|
|
3MBP
 
 | | MALTODEXTRIN-BINDING PROTEIN WITH BOUND MALTOTRIOSE | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Spurlino, J.C, Quiocho, F.A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor.
Structure, 5, 1997
|
|
4NPP
 
 | | The GLIC-His10 wild-type structure in equilibrium between the open and locally-closed (LC) forms | | Descriptor: | NICKEL (II) ION, Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CV7
 
 | | Crystal structure of Rhodococcus equi VapB | | Descriptor: | COBALT (II) ION, VIRULENCE ASSOCIATED PROTEIN VAPB | | Authors: | Geerds, C, Niemann, H.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Rhodococcus Equi Virulence-Associated Protein B (Vapb) Reveals an Eight-Stranded Antiparallel [Beta]-Barrel Consisting of Two Greek-Key Motifs
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4NQX
 
 | |
4FM9
 
 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
4NTC
 
 | | Crystal structure of GliT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GliT | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3MHR
 
 | | 14-3-3 sigma in complex with YAP pS127-peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schumacher, B, Skwarczynska, M, Rose, R, Ottmann, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of a 14-3-3[sigma]-YAP phosphopeptide complex at 1.15 A resolution
Acta Crystallogr.,Sect.F, 66, 2010
|
|
