2ARL
 
 | | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding | | Descriptor: | ACETIC ACID, CHLORIDE ION, GFP-like non-fluorescent chromoprotein, ... | | Authors: | Wilmann, P.G, Battad, J, Beddoe, T, Olsen, S, Smith, S.C, Dove, S, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2005-08-19 | | Release date: | 2006-09-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 angstroms crystal structure of a pocilloporin at pH 3.5: the structural basis for the linkage between color transition and halide binding
Photochem.Photobiol., 82, 2006
|
|
4ISU
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | Descriptor: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
2CEN
 
 | | P1' Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold | | Descriptor: | POL PROTEIN, {(1S)-1-[N'-[(2S)-2-HYDROXY-2-((1S,2R)-2-HYDROXY-INDAN-1-YLCARBAMOYL)-3-PHENYL-PROPYL]-N'-[4-(PYRIDINE-2-YL)-BENZYL]-HYDRAZINOCARBONYL]-2,2-DIMETHYL-PROPYL}-CARBAMIC ACID METHYL ESTER | | Authors: | Ginman, N, Ekegren, J.K, Johansson, A, Wallberg, H, Larhed, M, Samuelsson, B, Hallberg, A, Unge, T. | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Microwave-Accelerated Synthesis of P1'-Extended HIV-1 Protease Inhibitors Encompassing a Tertiary Alcohol in the Transition-State Mimicking Scaffold.
J.Med.Chem., 49, 2006
|
|
4KDS
 
 | |
4KYN
 
 | |
4JHI
 
 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-benzyladenine | | Descriptor: | MtN13 protein, N-BENZYL-9H-PURIN-6-AMINE, SODIUM ION | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2AC4
 
 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2CCR
 
 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, TRIETHYLENE GLYCOL, YVFO, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
4LBV
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Elongation factor Tu-A, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2GM2
 
 | | NMR structure of Xanthomonas campestris XCC1710: Northeast Structural Genomics Consortium target XcR35 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Xiao, R, Wang, D.Y, Ma, L.C, Ciano, M, Montelione, G.T, Ramelot, T.A, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Xanthomonas campestris XCC1710 protein
To be Published
|
|
4HOD
 
 | | Crystal structure of LeuT-E290S with bound Cl | | Descriptor: | CHLORIDE ION, LEUCINE, SODIUM ION, ... | | Authors: | Kantcheva, A.K, Quick, M, Shi, L, Winther, A.M.L, Stolzenberg, S, Weinstein, H, Javitch, J.A, Nissen, P. | | Deposit date: | 2012-10-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The chloride binding site of Neurotransmitter Sodium Symporters
Proc.Natl.Acad.Sci.USA, 2013
|
|
2AIX
 
 | |
4HW5
 
 | |
4LBW
 
 | | Identifying ligand binding hot spots in proteins using brominated fragments | | Descriptor: | AMMONIUM ION, Elongation factor Tu-A, MAGNESIUM ION, ... | | Authors: | Groftehauge, M.K, Therkelsen, M, Taaning, R, Skrydstrup, T, Morth, J.P, Nissen, P. | | Deposit date: | 2013-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identifying ligand-binding hot spots in proteins using brominated fragments.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1CM2
 
 | | STRUCTURE OF HIS15ASP HPR AFTER HYDROLYSIS OF RINGED SPECIES. | | Descriptor: | HISTIDINE-CONTAINING PROTEIN | | Authors: | Napper, S, Delbaere, L.T.J, Waygood, E.B. | | Deposit date: | 1999-05-13 | | Release date: | 2000-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartyl replacement of the active site histidine in histidine-containing protein, HPr, of the Escherichia coli Phosphoenolpyruvate:Sugar phosphotransferase system can accept and donate a phosphoryl group. Spontaneous dephosphorylation of acyl-phosphate autocatalyzes an internal cyclization
J.Biol.Chem., 274, 1999
|
|
4E6A
 
 | | p38a-PIA23 complex | | Descriptor: | (2S)-2-methoxy-3-(octadecyloxy)propyl (1R,2R,3R,4S,6S)-2,3,4-trihydroxy-6-(2-methylpropoxy)cyclohexyl hydrogen (S)-phosphate, Mitogen-activated protein kinase 14 | | Authors: | Livnah, O, Tzarum, N, Eisenberg-Domovich, Y. | | Deposit date: | 2012-03-15 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Lipid Molecules Induce p38 alpha Activation via a Novel Molecular Switch.
J.Mol.Biol., 424, 2012
|
|
7PJK
 
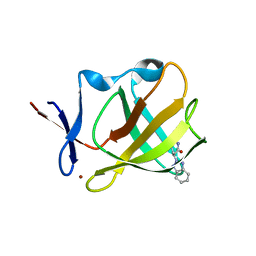 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with a benzotriazole analog of thalidomide | | Descriptor: | (3S)-3-(benzotriazol-2-yl)piperidine-2,6-dione, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Hartmann, M.D, Maiwald, S. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Replacing the phthalimide core in thalidomide with benzotriazole.
J Enzyme Inhib Med Chem, 37, 2022
|
|
4E5B
 
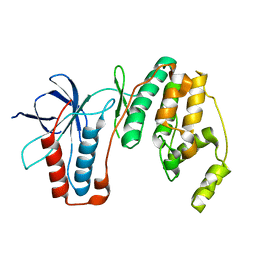 | |
4EH3
 
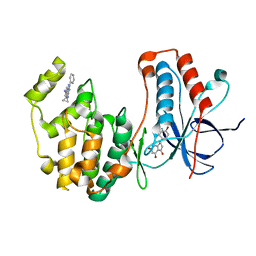 | | Human p38 MAP kinase in complex with NP-F2 and RL87 | | Descriptor: | Mitogen-activated protein kinase 14, NARINGENIN, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
5QYP
 
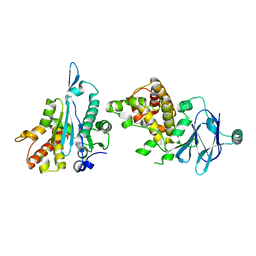 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 05 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYQ
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 06 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYU
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 10 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYV
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 11 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYW
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 12 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5QYZ
 
 | | PanDDA analysis group deposition -- Auto-refined data of Aar2/RNaseH for ground state model 15 | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
