6QW1
 
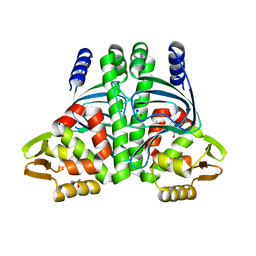 | | The Transcriptional Regulator PrfA-L140F mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
7ZY6
 
 | |
4YL0
 
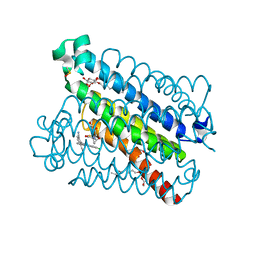 | | Crystal Structures of mPGES-1 Inhibitor Complexes | | Descriptor: | 2-(9-chloro-1H-phenanthro[9,10-d]imidazol-2-yl)benzene-1,3-dicarbonitrile, DI(HYDROXYETHYL)ETHER, GLUTATHIONE, ... | | Authors: | Luz, J.G, Antonysamy, S, Kuklish, S.L, Fisher, M.J. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics.
J.Med.Chem., 58, 2015
|
|
6QPC
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in nanodisc | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
5OE8
 
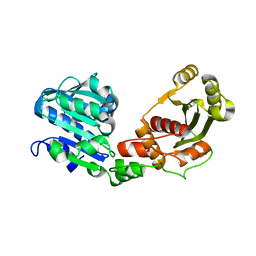 | | Structure of large terminase from the thermophilic bacteriophage D6E (Crystal form 2) | | Descriptor: | CHLORIDE ION, Large subunit terminase | | Authors: | Xu, R.G, Jenkins, H.T, Greive, S.J, Antson, A.A. | | Deposit date: | 2017-07-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the large terminase from a hyperthermophilic virus reveals a unique mechanism for oligomerization and ATP hydrolysis.
Nucleic Acids Res., 45, 2017
|
|
5OAR
 
 | | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Skerlova, J, Rezacova, P, Brynda, J, Pachl, P, Otwinowski, Z, Vanek, O. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae sheds light onto its substrate specificity, high stability, and regulation by propeptide.
FEBS J., 285, 2018
|
|
6QWG
 
 | | Serial Femtosecond Crystallography Structure of Cu Nitrite Reductase from Achromobacter cycloclastes: Nitrite complex at Room Temperature | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A, Moreno-Chicano, T, Appleby, M.V, Worrall, J.W, Duyvesteyn, H.M.E, Strange, R.W, Beale, J, Axford, D, Sherrell, D.A, Sugimoto, H, Owada, S, Tono, K, Owen, R.L. | | Deposit date: | 2019-03-05 | | Release date: | 2019-11-20 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
5LK5
 
 | |
7VB2
 
 | | Solution structure of human ribosomal protein uL11 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Lee, K.M, Wong, K.B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The flexible N-terminal motif of uL11 unique to eukaryotic ribosomes interacts with P-complex and facilitates protein translation.
Nucleic Acids Res., 50, 2022
|
|
6NFC
 
 | | BG505 MD64 N332-GT5 SOSIP trimer in complex with BG18-like precursor HMP42 fragmentantigen binding and base-binding RM20A3 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG18-like precursor HMP42 fragment antigen binding heavy chain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-19 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
7EA8
 
 | |
6T8D
 
 | | The cytotoxin MakB from Vibrio cholerae | | Descriptor: | MakB, SULFATE ION | | Authors: | Persson, K, Nagampalli, R. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A tripartite cytolytic toxin formed by Vibrio cholerae proteins with flagellum-facilitated secretion.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3ZM3
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
7SQF
 
 | |
7Z97
 
 | | Crystal structure of the F191M variant of Variovorax paradoxus indole monooxygenase (VpIndA1) in complex with 6-bromoindole | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kratky, J, Weisse, R, Strater, N. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Mechanistic Studies on Substrate and Stereoselectivity of the Indole Monooxygenase VpIndA1: New Avenues for Biocatalytic Epoxidations and Sulfoxidations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6DWF
 
 | |
5OEY
 
 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in holo form. | | Descriptor: | CITRIC ACID, FBP protein, MANGANESE (II) ION, ... | | Authors: | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
5OF8
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5I7O
 
 | |
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
3ZVA
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
6QPS
 
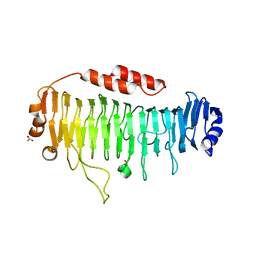 | | Structural characterization of a mannuronic acid specific polysaccharide family 6 lyase enzyme from human gut microbiota | | Descriptor: | ACETATE ION, CALCIUM ION, Polysaccharide Lyase Family 6 | | Authors: | Fredslund, F, Stender, E.G.P, Svensson, B, Welner, D.H. | | Deposit date: | 2019-02-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.287 Å) | | Cite: | Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbeBacteroides cellulosilyticus.
J.Biol.Chem., 294, 2019
|
|
5OFD
 
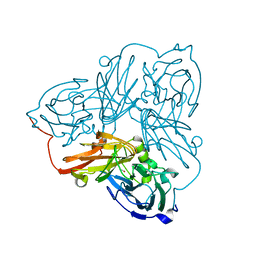 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
