2YAU
 
 | | X-ray structure of the Leishmania infantum tryopanothione reductase in complex with auranofin | | Descriptor: | 2,3,4,6-tetra-O-acetyl-1-thio-beta-D-glucopyranose, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G. | | Deposit date: | 2011-02-24 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A Gold-Containing Drug Against Parasitic Polyamine Metabolism: The X-Ray Structure of Trypanothione Reductase from Leishmania Infantum in Complex with Auranofin Reveals a Dual Mechanism of Enzyme Inhibition.
Amino Acids, 42, 2012
|
|
3NT1
 
 | | High resolution structure of naproxen:COX-2 complex. | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duggan, K.C, Musee, J, Walters, M.J, Harp, J.M, Kiefer, J.R, Oates, J.A, Marnett, L.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis for cyclooxygenase inhibition by the non-steroidal anti-inflammatory drug naproxen.
J.Biol.Chem., 285, 2010
|
|
1W4Q
 
 | | Binding of Nonnatural 3'-Nucleotides to Ribonuclease A | | Descriptor: | 2'-FLUORO-2'-DEOXYURIDINE 3'-MONOPHOSPHATE, PANCREATIC RIBONUCLEASE A | | Authors: | Jenkins, C.L, Thiyagarajan, N, Sweeney, R.Y, Guy, M.P, Kelemen, B.R, Acharya, K.R, Raines, R.T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Binding of Non-Natural 3'-Nucleotides to Ribonuclease A
FEBS J., 272, 2005
|
|
4J3P
 
 | | Crystal structure of full-length catechol oxidase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Hakulinen, N, Gasparetti, C, Kaljunen, H, Rouvinen, J. | | Deposit date: | 2013-02-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae.
J.Biol.Inorg.Chem., 18, 2013
|
|
6FX1
 
 | | Crystal structure of Pholiota squarrosa lectin in complex with an octasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, ... | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2A9C
 
 | | Crystal structure of R138Q mutant of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | GLYCEROL, MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A2A
 
 | |
2WU5
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhD His71Met mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of the E. Coli Succinate:Quinone Oxidoreductase (Sqr) Sdhd His71met Mutant
To be Published
|
|
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5GPN
 
 | | Architecture of mammalian respirasome | | Descriptor: | Acyl carrier protein, COPPER (II) ION, Cytochrome b, ... | | Authors: | Gu, J, Wu, M, Guo, R, Yang, M. | | Deposit date: | 2016-08-03 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | The architecture of the mammalian respirasome.
Nature, 537, 2016
|
|
3NT5
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9006 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
4H44
 
 | | 2.70 A Cytochrome b6f Complex Structure From Nostoc PCC 7120 | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Hasan, S.S, Yamashita, E, Baniulis, D, Cramer, W.A. | | Deposit date: | 2012-09-16 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2XAW
 
 | |
2Q6B
 
 | | Design and synthesis of novel, conformationally restricted HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[3-(4-FLUOROPHENYL)-1-ISOPROPYL-8-OXO-7-PHENYL-1,4,5,6,7,8-HEXAHYDROPYRROLO[2,3-C]AZEPIN-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-07-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of novel, conformationally restricted HMG-CoA reductase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2WLU
 
 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
3U5L
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a benzo-triazepine ligand (BzT-7) | | Descriptor: | 1,2-ETHANEDIOL, 8-chloro-1,4-dimethyl-6-phenyl-4H-[1,2,4]triazolo[4,3-a][1,3,4]benzotriazepine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Benzodiazepines and benzotriazepines as protein interaction inhibitors targeting bromodomains of the BET family.
Bioorg.Med.Chem., 20, 2012
|
|
3KHJ
 
 | |
1YD9
 
 | | 1.6A Crystal Structure of the Non-Histone Domain of the Histone Variant MacroH2A1.1. | | Descriptor: | Core histone macro-H2A.1, GOLD ION | | Authors: | Chakravarthy, S, Swamy, G.Y.S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the histone variant macroH2A
Mol.Cell.Biol., 25, 2005
|
|
2WZ6
 
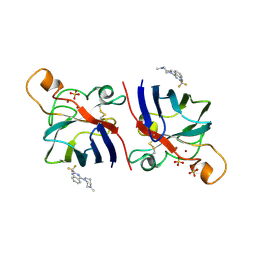 | | G93A SOD1 mutant complexed with Quinazoline. | | Descriptor: | 4-(4-METHYL-1,4-DIAZEPAN-1-YL)-2-(TRIFLUOROMETHYL)QUINAZOLINE, COPPER (II) ION, SULFATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2009-11-23 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Discovery of Small Molecule Binding Sites in Cu-Zn Human Superoxide Dismutase Familial Amyotrophic Lateral Sclerosis Mutants Provides Insights for Lead Optimization.
J.Med.Chem., 53, 2010
|
|
2FR0
 
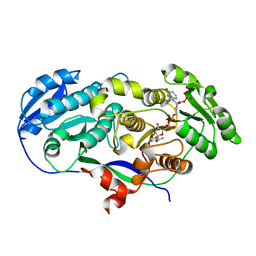 | |
1JR1
 
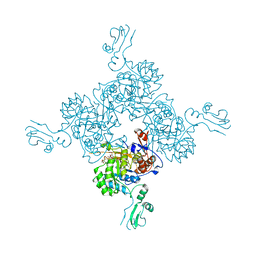 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
1F7T
 
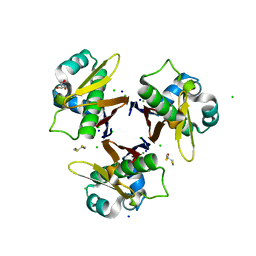 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE AT 1.8A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
6M6T
 
 | | Amylomaltase from Streptococcus agalactiae in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-1,5-anhydro-D-glucitol, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Wangkanont, K, Tumhom, S, Pongsawasdi, P. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Streptococcus agalactiae amylomaltase offers insight into the transglycosylation mechanism and the molecular basis of thermostability among amylomaltases.
Sci Rep, 11, 2021
|
|
1G6K
 
 | | Crystal structure of glucose dehydrogenase mutant E96A complexed with NAD+ | | Descriptor: | GLUCOSE 1-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamamoto, K, Kurisu, G, Kusunoki, M, Tabata, S, Urabe, I, Osaki, S. | | Deposit date: | 2000-11-06 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of stability-increasing mutants of glucose dehydrogenase
To be Published
|
|
4MQU
 
 | | Human GKRP complexed to AMG-3969 and S6P | | Descriptor: | 2-{4-[(2S)-4-[(6-aminopyridin-3-yl)sulfonyl]-2-(prop-1-yn-1-yl)piperazin-1-yl]phenyl}-1,1,1,3,3,3-hexafluoropropan-2-ol, D-SORBITOL-6-PHOSPHATE, GLYCEROL, ... | | Authors: | St Jean, D.J, Ashton, K.S, Bartberger, M.D, Chen, J, Chmait, S, Cupples, R, Galbreath, E, Helmering, J, Jordan, S.R, Liu, L. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Small molecule disruptors of the glucokinase-glucokinase regulatory protein interaction: 2. Leveraging structure-based drug design to identify analogues with improved pharmacokinetic profiles.
J.Med.Chem., 57, 2014
|
|
