2RCV
 
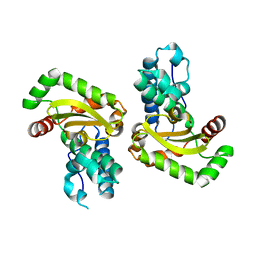 | | Crystal structure of the Bacillus subtilis superoxide dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Liu, P, Ewis, H.E, Huang, Y.J, Lu, C.D, Tai, P.C, Weber, I.T. | | Deposit date: | 2007-09-20 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis superoxide dismutase.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4CCY
 
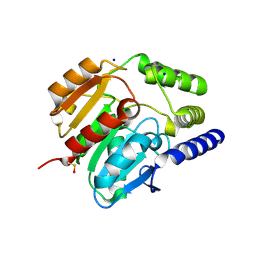 | | Crystal structure of carboxylesterase CesB (YbfK) from Bacillus subtilis | | Descriptor: | CARBOXYLESTERASE YBFK, SODIUM ION | | Authors: | Rozeboom, H.J, Godinho, L.F, Nardini, M, Quax, W.J, Dijkstra, B.W. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structures of Two Bacillus Carboxylesterases with Different Enantioselectivities.
Biochim.Biophys.Acta, 1844, 2014
|
|
4C74
 
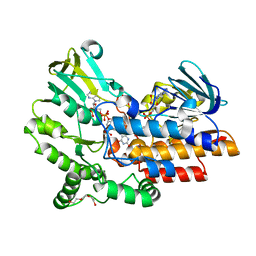 | | Phenylacetone monooxygenase: Reduced enzyme in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catal., 3, 2013
|
|
1SH7
 
 | |
2J4X
 
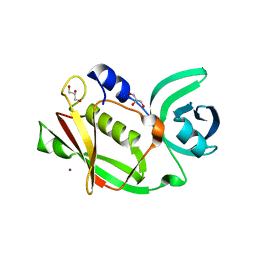 | | Streptococcus dysgalactiae-derived mitogen (SDM) | | Descriptor: | GLYCEROL, MITOGEN, ZINC ION | | Authors: | Saarinen, S, Kato, H, Uchiyama, T, Miyoshi-Akiyama, T, Papageorgiou, A.C. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Streptococcus Dysgalactiae-Derived Mitogen Reveals a Zinc-Binding Site and Alterations in Tcr Binding.
J.Mol.Biol., 373, 2007
|
|
1SJW
 
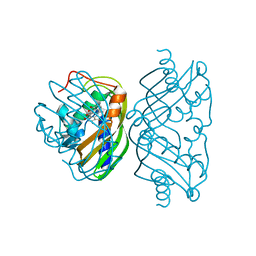 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
4D1Q
 
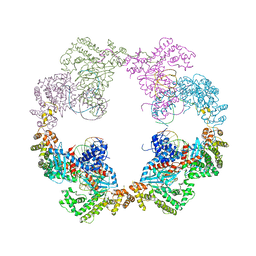 | | Hermes transposase bound to its terminal inverted repeat | | Descriptor: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | Authors: | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | Deposit date: | 2014-05-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
4C77
 
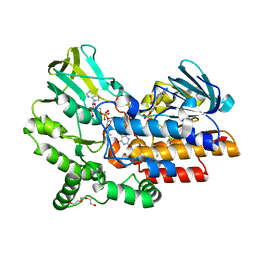 | | Phenylacetone monooxygenase: oxidised R337K mutant in complex with APADP | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, ... | | Authors: | Martinoli, C, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2013-09-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.
Acs Catal., 3, 2013
|
|
4D7E
 
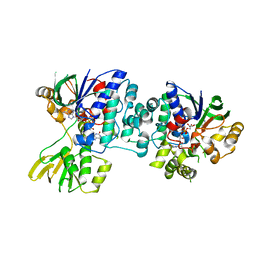 | | An unprecedented NADPH domain conformation in Lysine Monooxygenase NbtG from Nocardia farcinica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-LYS MONOOXYGENASE | | Authors: | Binda, C, Robinson, R, Keul, N, Rodriguez, P, Robinson, H.H, Mattevi, A, Sobrado, P. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An Unprecedented Nadph Domain Conformation in Lysine Monooxygenase Nbtg Provides Insights Into Uncoupling of Oxygen Consumption from Substrate Hydroxylation.
J.Biol.Chem., 290, 2015
|
|
4DMF
 
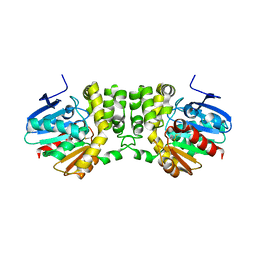 | |
4DMK
 
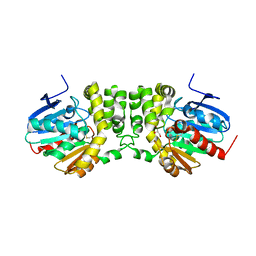 | |
2XAW
 
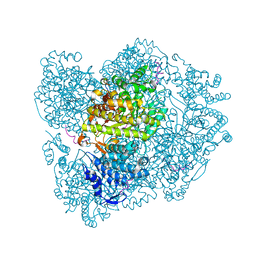 | |
4EX6
 
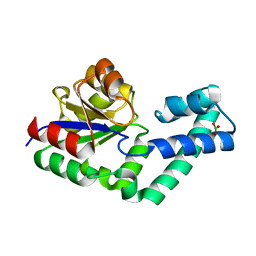 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
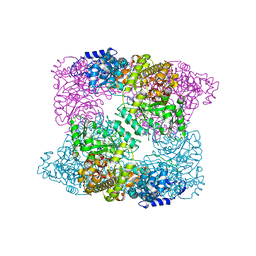
2XAY
 
 | |
2I3R
 
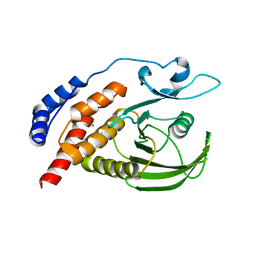 | | Engineered catalytic domain of protein tyrosine phosphatase HPTPbeta | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I3U
 
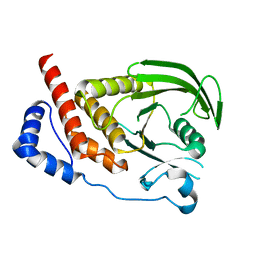 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-20 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I4E
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Receptor-type tyrosine-protein phosphatase beta, VANADATE ION | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4EHB
 
 | |
2I4G
 
 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with a sulfamic acid (soaking experiment) | | Descriptor: | CHLORIDE ION, N-(TERT-BUTOXYCARBONYL)-L-TYROSYL-N-METHYL-4-(SULFOAMINO)-L-PHENYLALANINAMIDE, Receptor-type tyrosine-protein phosphatase beta | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2I5X
 
 | | Engineering the PTPbeta catalytic domain with improved crystallization properties | | Descriptor: | (4-ETHYLPHENYL)SULFAMIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Klopfenstein, S. | | Deposit date: | 2006-08-26 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering the catalytic domain of human protein tyrosine phosphatase beta for structure-based drug discovery.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IPI
 
 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2VND
 
 | | The N69Q mutant of Vibrio cholerae endonuclease I | | Descriptor: | CHLORIDE ION, ENDONUCLEASE I, MAGNESIUM ION | | Authors: | Niiranen, L, Altermark, B, Brandsdal, B.O, Leiros, H.-K.S, Helland, R, Smalas, A.O, Willassen, N.P. | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effects of Salt on the Kinetics and Thermodynamic Stability of Endonuclease I from Vibrio Salmonicida and Vibrio Cholerae.
FEBS J., 275, 2008
|
|
1S2N
 
 | |
2XAZ
 
 | |
2J3K
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
