5DWQ
 
 | |
2RIM
 
 | | Crystal structure of Rtt109 | | Descriptor: | Regulator of Ty1 transposition protein 109 | | Authors: | Yuan, Y.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into histone h3 lysine 56 acetylation by rtt109
Structure, 16, 2008
|
|
2V85
 
 | |
2V88
 
 | |
2XB1
 
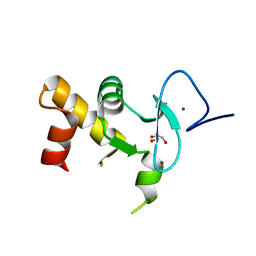 | | Crystal structure of the human Pygo2 PHD finger in complex with the B9L HD1 domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYGOPUS HOMOLOG 2, ... | | Authors: | Miller, T.C, Rutherford, T.J, Johnson, C.M, Fiedler, M, Bienz, M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Remodeling of the Histone H3 Binding Pocket in the Pygo2 Phd Finger Triggered by its Binding to the B9L/Bcl9 Co-Factor.
J.Mol.Biol., 401, 2010
|
|
7MJU
 
 | |
5DAH
 
 | |
2V86
 
 | |
5XM2
 
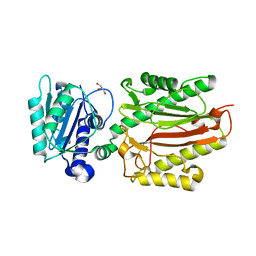 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
2DEX
 
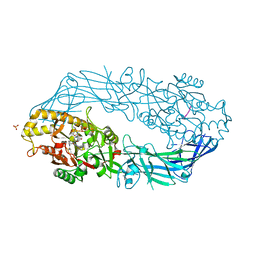 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal peptide including Arg17 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2DEW
 
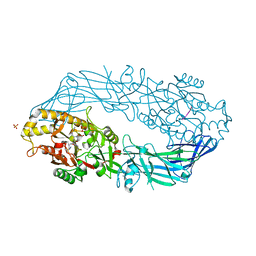 | | Crystal structure of human peptidylarginine deiminase 4 in complex with histone H3 N-terminal tail including Arg8 | | Descriptor: | 10-mer peptide from histone H3, CALCIUM ION, Protein-arginine deiminase type IV, ... | | Authors: | Arita, K, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Sato, M. | | Deposit date: | 2006-02-18 | | Release date: | 2006-04-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4YS3
 
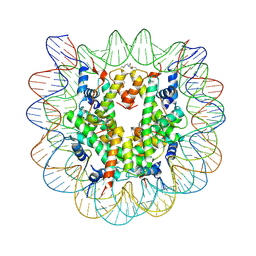 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
5H6Q
 
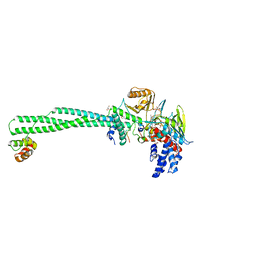 | | Crystal structure of LSD1-CoREST in complex with peptide 11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5H6R
 
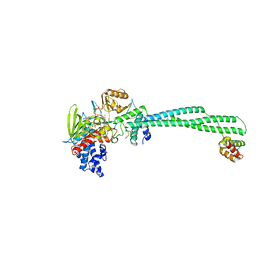 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
4Z66
 
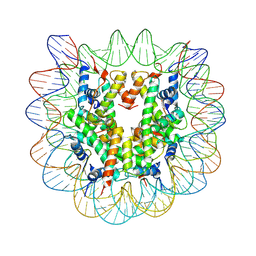 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-04-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
2L5A
 
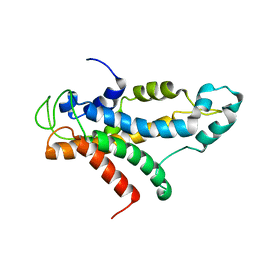 | | Structural basis for recognition of centromere specific histone H3 variant by nonhistone Scm3 | | Descriptor: | Histone H3-like centromeric protein CSE4, Protein SCM3, Histone H4 | | Authors: | Zhou, Z, Feng, H, Zhou, B, Ghirlando, R, Hu, K, Zwolak, A, Jenkins, L, Xiao, H, Tjandra, N, Wu, C, Bai, Y. | | Deposit date: | 2010-10-28 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3.
Nature, 472, 2011
|
|
4K3D
 
 | | Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
3DB3
 
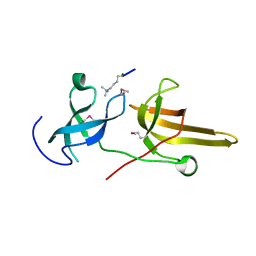 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3U5O
 
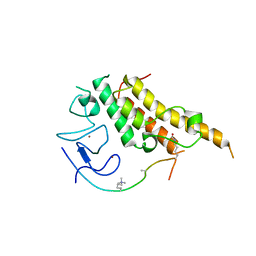 | |
2RVL
 
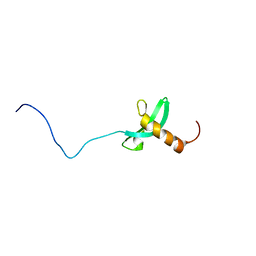 | |
3PNW
 
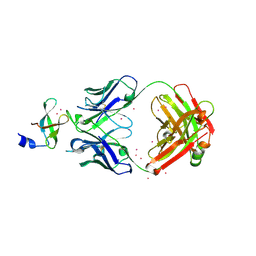 | | Crystal Structure of the tudor domain of human TDRD3 in complex with an anti-TDRD3 FAB | | Descriptor: | FAB heavy chain, FAB light chain, Tudor domain-containing protein 3, ... | | Authors: | Loppnau, P, Tempel, W, Wernimont, A.K, Lam, R, Ravichandran, M, Adams-Cioaba, M.A, Persson, H, Sidhu, S.S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CDR-H3 Diversity Is Not Required for Antigen Recognition by Synthetic Antibodies.
J.Mol.Biol., 425, 2013
|
|
3U5P
 
 | |
2AJ3
 
 | | Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 | | Descriptor: | Fab m18, Heavy Chain, Light Chain, ... | | Authors: | Prabakaran, P, Gan, J, Wu, Y.Q, Zhang, M.Y, Dimitrov, D.S, Ji, X. | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural mimicry of CD4 by a cross-reactive HIV-1 neutralizing antibody with CDR-H2 and H3 containing unique motifs.
J.Mol.Biol., 357, 2006
|
|
