4YDI
 
 | | Crystal structure of broad and potently neutralizing VRC01-class antibody Z258-VRC27.01, isolated from human donor Z258, in complex with HIV-1 gp120 from clade A strain Q23.17 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.452 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
6GMC
 
 | | 1.2 A resolution structure of human hydroxyacid oxidase 1 bound with FMN and 4-carboxy-5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, ... | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Smee, C, Arrowsmith, C.H, Edwards, E, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate
To Be Published
|
|
8W2J
 
 | | Human liver phosphofructokinase-1 filament in the T-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
8W2H
 
 | | Human liver phosphofructokinase-1 in the T-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
8W2G
 
 | | Human liver phosphofructokinase-1 in the R-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
8W2I
 
 | | Human liver phosphofructokinase-1 filament in the R-state conformation | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B.A. | | Deposit date: | 2024-02-20 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for allosteric regulation of human phosphofructokinase-1.
Nat Commun, 15, 2024
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
7MHV
 
 | |
8VUF
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
6P1X
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and L-ddCTP | | Descriptor: | DNA Primer 20-mer, DNA template 27-mer, MAGNESIUM ION, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-20 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
5NEE
 
 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor 5-[2-(morpholine-4-carbonyl)1,3-oxazol-5-yl)]thiophene-2-sulfonammide | | Descriptor: | 5-(2-morpholin-4-ylcarbonyl-1,3-oxazol-5-yl)thiophene-2-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Krasavin, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1,3-Oxazole-based selective picomolar inhibitors of cytosolic human carbonic anhydrase II alleviate ocular hypertension in rabbits: Potency is supported by X-ray crystallography of two leads.
Bioorg. Med. Chem., 25, 2017
|
|
8W0K
 
 | |
8CBR
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBU
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT884 | | Descriptor: | (2S)-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2H-chromen-6-yl)phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBT
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT872 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
6GH3
 
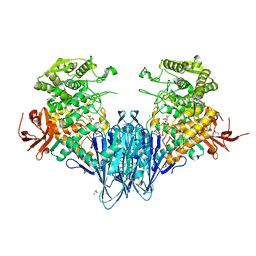 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-man-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
5MAR
 
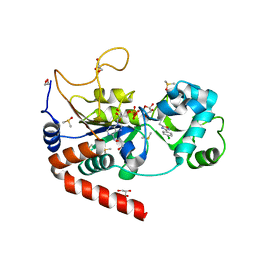 | | Structure of human SIRT2 in complex with 1,2,4-Oxadiazole inhibitor and ADP ribose. | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(4-chlorophenyl)-1,2,4-oxadiazol-5-yl]propan-1-ol, ACETATE ION, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Development of 1,2,4-Oxadiazoles as Potent and Selective Inhibitors of the Human Deacetylase Sirtuin 2: Structure-Activity Relationship, X-ray Crystal Structure, and Anticancer Activity.
J. Med. Chem., 60, 2017
|
|
3EOJ
 
 | | Fmo protein from Prosthecochloris Aestuarii 2K AT 1.3A Resolution | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tronrud, D.E, Wen, J, Gay, L, Blankenship, R.E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
2WAJ
 
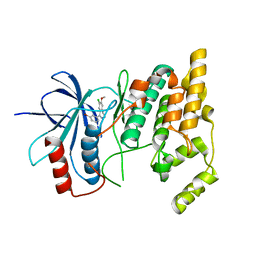 | | Crystal structure of human Jnk3 complexed with a 1-aryl-3,4- dihydroisoquinoline inhibitor | | Descriptor: | 1-(3-BROMOPHENYL)-7-CHLORO-6-METHOXY-3,4-DIHYDROISOQUINOLINE, MITOGEN-ACTIVATED PROTEIN KINASE 10 | | Authors: | Bax, B.D, Christopher, J.A, Jones, E.J, Mosley, J.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1-Aryl-3,4-Dihydroisoquinoline Inhibitors of Jnk3.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6CQA
 
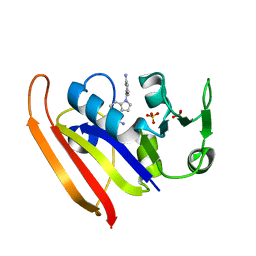 | | E. coli DHFR complex with inhibitor AMPQD | | Descriptor: | 7-[(3-aminophenyl)methyl]-7H-pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cao, H, Rodrigues, J, Benach, J, Wasserman, S, Morisco, L, Koss, J, Shakhnovich, E, Skolnick, J. | | Deposit date: | 2018-03-14 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a tetrahydrofolate-bound dihydrofolate reductase reveals the origin of slow product release.
Commun Biol, 1, 2018
|
|
6GH2
 
 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-glc-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
3FB1
 
 | |
5NJ1
 
 | |
5YFX
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase mutant D204N from Pyrococcus horikoshii OT3 in complex with ribose-1,5-bisphosphate and AMP | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Ribose 1,5-bisphosphate isomerase | | Authors: | Gogoi, P, Kanaujia, S.P. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3.
Sci Rep, 8, 2018
|
|
