4JNA
 
 | | Crystal structure of the DepH complex with dimethyl-FK228 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DepH, ... | | Authors: | Li, J, Wang, C, Zhang, Z.M, Zhou, J.H, Cheng, E. | | Deposit date: | 2013-03-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis.
Sci Rep, 4, 2014
|
|
4FW8
 
 | | Crystal structure of FABG4 complexed with Coenzyme NADH | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2012-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
3W1M
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-bromoorotate | | Descriptor: | 5-bromo-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A. | | Deposit date: | 2012-11-16 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-bromoorotate
To be Published
|
|
3W2J
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-135 | | Descriptor: | 5-[2-(3,4-difluorophenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-135
To be Published
|
|
8SDB
 
 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
5NZI
 
 | | Crystal structure of UDP-glucose pyrophosphorylase S374F mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5XUU
 
 | | Crystal structure of Lachnospiraceae bacterium ND2006 Cpf1 in complex with crRNA and target DNA (TCCA PAM) | | Descriptor: | 1,2-ETHANEDIOL, DNA (29-MER), DNA (5'-D(*CP*GP*TP*CP*CP*TP*CP*CP*A)-3'), ... | | Authors: | Yamano, T, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2017-06-26 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Canonical and Non-canonical PAM Recognition by CRISPR-Cpf1.
Mol. Cell, 67, 2017
|
|
4AWS
 
 | |
3W1T
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-095 | | Descriptor: | 2,6-dioxo-5-{2-[3-(trifluoromethyl)phenyl]ethyl}-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-21 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-095
To be Published
|
|
5FPP
 
 | | Structure of a pre-reaction ternary complex between sarin- acetylcholinesterase and HI-6 | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9IHW
 
 | |
9II0
 
 | |
1V3V
 
 | | Crystal structure of leukotriene B4 12-hydroxydehydrogenase/15-oxo-prostaglandin 13-reductase complexed with NADP and 15-oxo-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hori, T, Yokomizo, T, Ago, H, Sugahara, M, Ueno, G, Yamamoto, M, Kumasaka, T, Shimizu, T, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-06 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of leukotriene B4 12-hydroxydehydrogenase/15-Oxo-prostaglandin 13-reductase catalytic mechanism and a possible Src homology 3 domain binding loop
J.Biol.Chem., 279, 2004
|
|
3W22
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125 | | Descriptor: | 5-[2-(4-tert-butylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-125
To be Published
|
|
3VTE
 
 | | Crystal structure of tetrahydrocannabinolic acid synthase from Cannabis sativa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Tetrahydrocannabinolic acid synthase | | Authors: | Shoyama, Y, Tamada, T, Kurihara, K, Takeuchi, A, Taura, F, Arai, S, Blaber, M, Shoyama, Y, Morimoto, S, Kuroki, R. | | Deposit date: | 2012-05-28 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa
J.Mol.Biol., 423, 2012
|
|
3W1U
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-111 | | Descriptor: | 5-[2-(4-methylphenyl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-21 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-3-111
To be Published
|
|
5W8J
 
 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | Descriptor: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lukacs, C.M, Moulin, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
2V8T
 
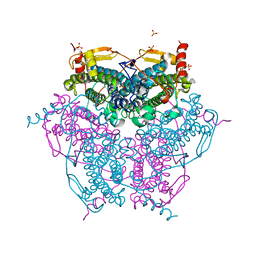 | | Crystal structure of Mn catalase from Thermus Thermophilus complexed with chloride | | Descriptor: | CHLORIDE ION, LITHIUM ION, MANGANESE (II) ION, ... | | Authors: | Antonyuk, S.V, Barynin, V.V, Vaguine, A.A, Melik-Adamyan, W.R, Popov, A.N, Lamsin, V.S, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 2007-08-14 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Three-Dimentional Structure of the Enzyme Dimanganese Catalase from Thermus Thermophilus at 1 Angstrom Resolution
Crystallogr.Rep.(Transl. Kristallografiya), 45, 2000
|
|
6UNI
 
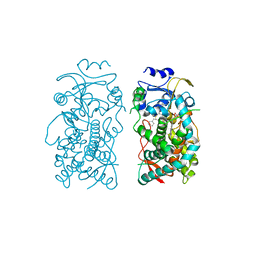 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(1H-indol-3-yl)-3-{[(2R)-1-oxo-3-phenyl-1-{[2-(pyridin-3-yl)ethyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
5NZL
 
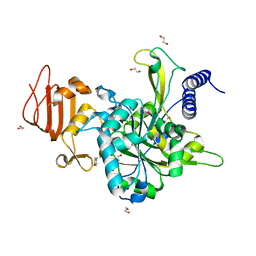 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with resveratrol | | Descriptor: | 1,2-ETHANEDIOL, RESVERATROL, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
5TI2
 
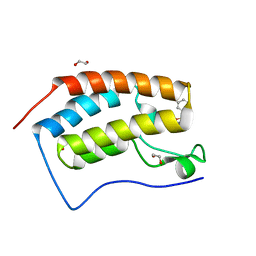 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 7635936 | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5OLU
 
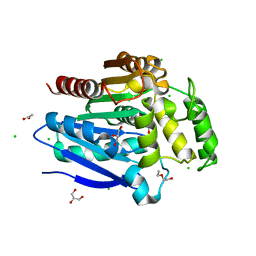 | | The crystal structure of a highly thermostable carboxyl esterase from Bacillus coagulans in complex with glycerol | | Descriptor: | ACETATE ION, Alpha/beta hydrolase family protein, CHLORIDE ION, ... | | Authors: | Gourlay, L.J, Nakhnoukh, C, Bolognesi, M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold.
FEBS J., 285, 2018
|
|
1CJC
 
 | |
5TI3
 
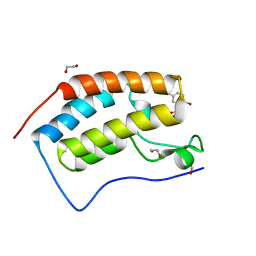 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 17503468 | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dibromo-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5Y7X
 
 | | Human Peroxisome proliferator-activated receptor (PPAR) delta in complexed with a potent and selective agonist | | Descriptor: | 2-[2-methyl-4-[[4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-selenazol-5-yl]methylsulfanyl]phenoxy]ethanoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Kim, H.L, Chin, J.W, Cho, S.J, Song, J.Y, Yoon, H.S, Bae, J.H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Highly potent and selective PPAR delta agonist reverses memory deficits in mouse models of Alzheimer's disease.
Theranostics, 14, 2024
|
|
