5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z87
 
 | | Structural of a novel b-glucosidase EmGH1 at 2.3 angstrom from Erythrobacter marinus | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BENZAMIDINE, ... | | Authors: | Li, J.X, Hu, X.J, Zhao, Y, Li, L. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analysis of a novel b-glucosidase EmGH1 from Erythrobacter marinus
To Be Published
|
|
7E6I
 
 | | Glucose-6-phosphate dehydrogenase in complex with its substrate glucose-6-phosphate | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Vu, H.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
5TT1
 
 | |
2VN0
 
 | | CYP2C8DH COMPLEXED WITH TROGLITAZONE | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, CYTOCHROME P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J. Biol. Chem., 283, 2008
|
|
5ICF
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
6CM4
 
 | | Structure of the D2 Dopamine Receptor Bound to the Atypical Antipsychotic Drug Risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, D(2) dopamine receptor, endolysin chimera, ... | | Authors: | Wang, S, Che, T, Levit, A, Shoichet, B.K, Wacker, D, Roth, B.L. | | Deposit date: | 2018-03-02 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone.
Nature, 555, 2018
|
|
5IHH
 
 | | Crystal structure of human transthyretin in complex with luteolin-MeO at 1.35 A resolution | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5-hydroxy-7-methoxy-4H-1-chromen-4-one, SODIUM ION, Transthyretin | | Authors: | Begum, A, Nilsson, L, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity.
Plos One, 11, 2016
|
|
2X3U
 
 | | Ferredoxin-NADP reductase mutant with Tyr 303 replaced by Phe (Y303F) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Peregrina, J.R, Sanchez-Azqueta, A, Medina, M. | | Deposit date: | 2010-01-27 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of Specific Residues in Coenzyme Binding, Charge-Transfer Complex Formation, and Catalysis in Anabaena Ferredoxin Nadp(+)-Reductase.
Biochim.Biophys.Acta, 1797, 2010
|
|
4H4C
 
 | | IspH in complex with (E)-4-fluoro-3-methylbut-2-enyl diphosphate | | Descriptor: | (2E)-4-fluoro-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Eisenreich, W, Jauch, J, Bacher, A, Groll, M. | | Deposit date: | 2012-09-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Fluoro, Amino, and Thiol Inhibitors Bound to the [Fe(4) S(4) ] Protein IspH.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5Z2L
 
 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
4QNW
 
 | |
5UUT
 
 | | N-myristoyltransferase 1 (NMT) bound to myristoyl-CoA | | Descriptor: | CITRIC ACID, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Goodwin, O, Pegan, S. | | Deposit date: | 2017-02-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Blocking Myristoylation of Src Inhibits Its Kinase Activity and Suppresses Prostate Cancer Progression.
Cancer Res., 77, 2017
|
|
7M8V
 
 | | Human CYP11B2 in complex with LCI699 | | Descriptor: | 4-[(4R,5R)-6,7-dihydro-5H-pyrrolo[1,2-c]imidazol-5-yl]-3-fluorobenzonitrile, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Aldosterone Synthase Structure With Cushing Disease Drug LCI699 Highlights Avenues for Selective CYP11B Drug Design.
Hypertension, 78, 2021
|
|
2XPD
 
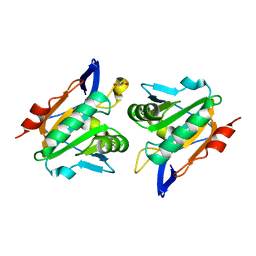 | | Reduced Thiol peroxidase (Tpx) from yersinia Pseudotuberculosis | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, THIOL PEROXIDASE | | Authors: | Gabrielsen, M, Zetterstrom, C.E, Wang, D, Elofsson, M, Roe, A.J. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
3ZV4
 
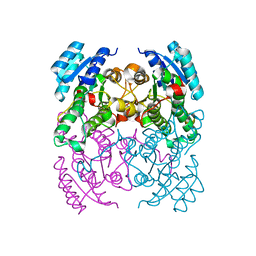 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 IN APO FORM AT 1.8 ANGSTROM | | Descriptor: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | Authors: | Dhindwal, S, Patil, D.N, Kumar, P. | | Deposit date: | 2011-07-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
2ZUP
 
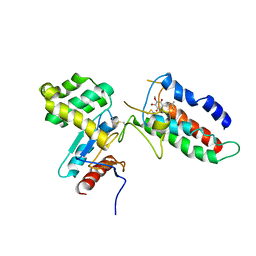 | | Updated crystal structure of DsbB-DsbA complex from E. coli | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S, Nakagawa, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
4DU8
 
 | |
4DDD
 
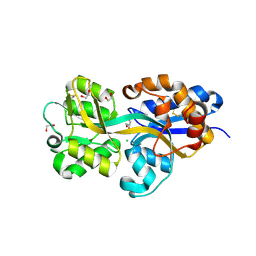 | |
4PW3
 
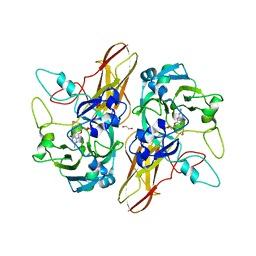 | |
4I9M
 
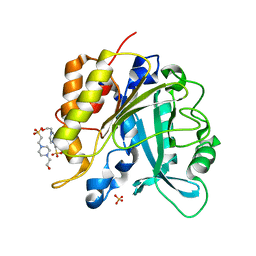 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
2QT5
 
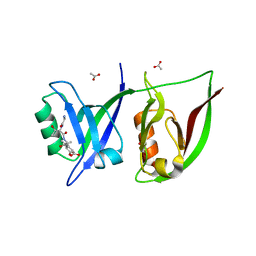 | | Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide | | Descriptor: | (ASN)(ASN)(LEU)(GLN)(ASP)(GLY)(THR)(GLU)(VAL), 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Long, J, Wei, Z, Feng, W, Zhao, Y, Zhang, M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
J.Mol.Biol., 375, 2008
|
|
1T4D
 
 | | Crystal structure of Escherichia coli aspartate beta-semialdehyde dehydrogenase (EcASADH), at 1.95 Angstrom resolution | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
3COX
 
 | | CRYSTAL STRUCTURE OF CHOLESTEROL OXIDASE COMPLEXED WITH A STEROID SUBSTRATE. IMPLICATIONS FOR FAD DEPENDENT ALCOHOL OXIDASES | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Vrielink, A, Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1993-06-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases.
Biochemistry, 32, 1993
|
|
5W07
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-30 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor of Mycobacterium tuberculosis InhA
To Be Published
|
|
