6UY7
 
 | |
9ERX
 
 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
8SXU
 
 | |
8SXT
 
 | | Structure of LINE-1 ORF2p with template:primer hybrid | | Descriptor: | DNA primer, LINE-1 retrotransposable element ORF2 protein, MAGNESIUM ION, ... | | Authors: | van Eeuwen, T, Taylor, M.S, Rout, M.P. | | Deposit date: | 2023-05-24 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
4UFN
 
 | | Laboratory evolved variant R-C1B1 of potato epoxide hydrolase StEH1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EPOXIDE HYDROLASE | | Authors: | Carlsson, A.J, Bauer, P, Nilsson, M, Dobritzsch, D, Kamerlin, S.C.L, Widersten, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Diversity and Enantioconvergence in Potato Epoxide Hydrolase 1.
Org.Biomol.Chem., 14, 2016
|
|
7Z1V
 
 | | PARP15 catalytic domain in complex with OUL208 | | Descriptor: | 6-methoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z1Y
 
 | | PARP15 catalytic domain in complex with OUL245 | | Descriptor: | DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15, [1,2,4]triazolo[3,4-b][1,3]benzothiazol-6-ol | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
7Z63
 
 | |
7Z1W
 
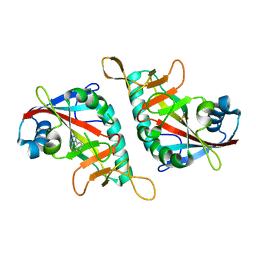 | | PARP15 catalytic domain in complex with OUL246 | | Descriptor: | 6-chloranyl-[1,2,4]triazolo[3,4-b][1,3]benzothiazole, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
4D4B
 
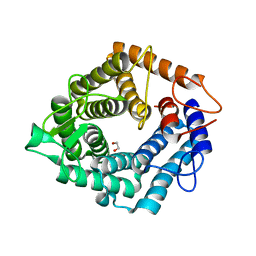 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7KLC
 
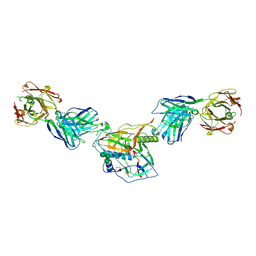 | |
4XHU
 
 | |
6UIP
 
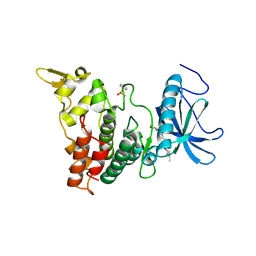 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
8SU8
 
 | |
8T09
 
 | |
8T7V
 
 | |
6UUI
 
 | |
8TCV
 
 | | Structure of PYCR1 complexed with 4-bromobenzene-1,3-dicarboxylic acid | | Descriptor: | 4-bromobenzene-1,3-dicarboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TCZ
 
 | |
8TCX
 
 | | Structure of PYCR1 complexed with 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydroquinazoline-6-carboxylic acid, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
8TCW
 
 | |
8TCU
 
 | |
8TCY
 
 | | Structure of PYCR1 complexed with 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid | | Descriptor: | 7-fluoro-2-oxo-1,2,3,4-tetrahydroquinoline-6-carboxylic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Fragment Inhibitors of PYCR1 from Docking-Guided X-ray Crystallography.
J.Chem.Inf.Model., 64, 2024
|
|
7Z2Q
 
 | | PARP15 catalytic domain in complex with OUL232 | | Descriptor: | 5,8-dimethoxy-[1,2,4]triazolo[3,4-b][1,3]benzothiazol-1-amine, DIMETHYL SULFOXIDE, Protein mono-ADP-ribosyltransferase PARP15 | | Authors: | Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2022-02-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | [1,2,4]Triazolo[3,4- b ]benzothiazole Scaffold as Versatile Nicotinamide Mimic Allowing Nanomolar Inhibition of Different PARP Enzymes.
J.Med.Chem., 66, 2023
|
|
4XII
 
 | | X-ray structure of human butyrylcholinesterase in complex with N-((1-(2,3-dihydro-1H-inden-2-yl)piperidin-3-yl)methyl)-8-hydroxy-N-(2-methoxyethyl)-5-nitroquinoline-7-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Knez, D, Boris, B, Coquelle, N, Sosic, I, Sink, R, Brazzolotto, X, Mravljak, J, Colletier, J.P, Gobec, S. | | Deposit date: | 2015-01-07 | | Release date: | 2015-07-08 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based development of nitroxoline derivatives as potential multifunctional anti-Alzheimer agents.
Bioorg.Med.Chem., 23, 2015
|
|
