6SQR
 
 | | Crystal structure of Cat MDM2-S429E RING domain bound to UbcH5B-Ub | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
9GWV
 
 | |
6H6M
 
 | |
9GRN
 
 | | Crystal structure of the engineered C-terminal phosphatase domain from Saccharomyces cerevisiae Vip1 (apo, loop deletion residues 848-918) | | Descriptor: | ACETATE ION, GLYCEROL, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Raia, P, Lee, K, Hothorn, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A small signaling domain controls PPIP5K phosphatase activity in phosphate homeostasis.
Nat Commun, 16, 2025
|
|
6T60
 
 | |
6GMR
 
 | | pVHL:EloB:EloC in complex with (4-(1H-pyrrol-1-yl)phenyl)methanol | | Descriptor: | (4-pyrrol-1-ylphenyl)methanol, Elongin-B, Elongin-C, ... | | Authors: | Van Molle, I, Lucas, X, Ciulli, A. | | Deposit date: | 2018-05-28 | | Release date: | 2018-08-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
6E53
 
 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
6E5G
 
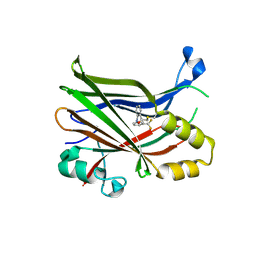 | |
6GVZ
 
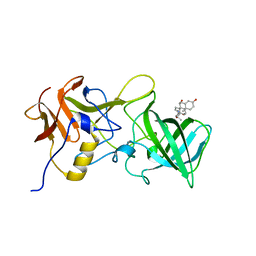 | |
6HIV
 
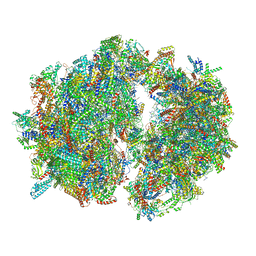 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete mitoribosome | | Descriptor: | 12S rRNA, 50S ribosomal protein L13, putative, ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boerhringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
4Y88
 
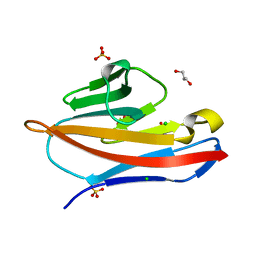 | |
8UZE
 
 | | Crystal structure of chimeric bat coronavirus BANAL-20-236 RBD complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for mouse receptor recognition by bat SARS2-like coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1N4V
 
 | |
5NAJ
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH | | Descriptor: | (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{R},7~{S},10~{S},13~{R})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, (3~{S},7~{R},10~{R},13~{S})-4-[[(3~{S},7~{R},10~{R},13~{S})-4-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-en-13-yl]carbonyl]-2-oxidanylidene-1,4-diazatricyclo[8.3.0.0^{3,7}]tridec-8-ene-13-carboxylic acid, CHLORIDE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TGW
 
 | |
3II4
 
 | |
9EVK
 
 | | Crystal Structure of human Collagen Hydroxylysine Galactosyltransferase GLT25D1/COLGALT1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | De Marco, M, Scietti, L, Rai, S.R, Mattoteia, D, Pinnola, A, Forneris, F. | | Deposit date: | 2024-03-30 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1.
Nat Commun, 16, 2025
|
|
9EVJ
 
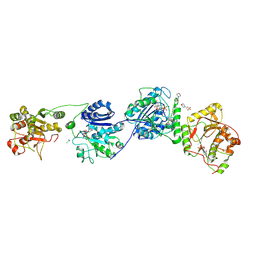 | | Crystal Structure of human Collagen Hydroxylysine Galactosyltransferase GLT25D1/COLGALT1: complex with Mn2+ and UDP-Gal | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | De Marco, M, Scietti, L, Rai, S.R, Mattoteia, D, Pinnola, A, Forneris, F. | | Deposit date: | 2024-03-30 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1.
Nat Commun, 16, 2025
|
|
9EVL
 
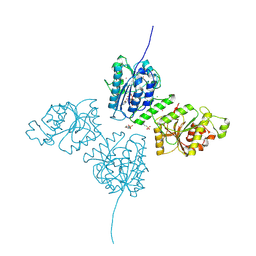 | | Crystal Structure of human Collagen Hydroxylysine Galactosyltransferase GLT25D1/COLGALT1: Hg2+ soak for experimental phasing | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | De Marco, M, Scietti, L, Rai, S.R, Mattoteia, D, Pinnola, A, Forneris, F. | | Deposit date: | 2024-03-30 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure and enzymatic mechanism of the human collagen hydroxylysine galactosyltransferase GLT25D1/COLGALT1.
Nat Commun, 16, 2025
|
|
8JE0
 
 | | A novel amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, Amidase, ZINC ION | | Authors: | Ma, D, Feng, R. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel amidohydrolase catalyze the degradation of PAM by Klebsiella sp. PCX
To Be Published
|
|
9BQE
 
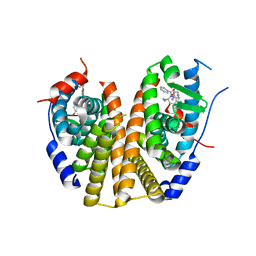 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-(2-(1-propylpyrrolidin-3-yl)ethoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-(4-{[(3R)-1-propylpyrrolidin-3-yl]methoxy}phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Young, K.Y, Fanning, S.W. | | Deposit date: | 2024-05-09 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting unique ligand binding domain structural features downregulates DKK1 in Y537S ESR1 mutant breast cancer cells.
Breast Cancer Res, 27, 2025
|
|
9BU1
 
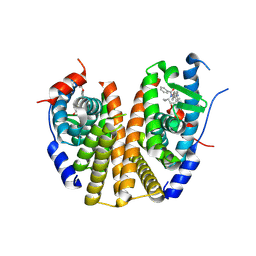 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with (6'-hydroxy-1'-(4-((1-propylazetidin-3-yl)methoxy)phenyl)-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl)(phenyl)methanone | | Descriptor: | Estrogen receptor, [(1'R)-6'-hydroxy-1'-{4-[(1-propylazetidin-3-yl)methoxy]phenyl}-1',4'-dihydro-2'H-spiro[cyclopropane-1,3'-isoquinolin]-2'-yl](phenyl)methanone | | Authors: | Young, K.S, Fanning, S.W. | | Deposit date: | 2024-05-16 | | Release date: | 2024-06-19 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Targeting unique ligand binding domain structural features downregulates DKK1 in Y537S ESR1 mutant breast cancer cells.
Breast Cancer Res, 27, 2025
|
|
5D6M
 
 | | Mn(II)-loaded MnCcP.1 | | Descriptor: | Cytochrome c peroxidase, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Robinson, H, Gao, Y.-G, Hosseinzadeh, P, Lu, Y. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Enhancing Mn(II)-Binding and Manganese Peroxidase Activity in a Designed Cytochrome c Peroxidase through Fine-Tuning Secondary-Sphere Interactions.
Biochemistry, 55, 2016
|
|
7Y0R
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, 4-METHYLANILINE, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87L/V78S/A184V in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-toluidine and hydroxylamine
To Be Published
|
|
5A5W
 
 | | Crystal structure of Salmonella enterica HisA D7N D176A with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Soderholm, A, Guo, X, Newton, M.S, Evans, G.B, Nasvall, J, Patrick, W.M, Selmer, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-Step Ligand Binding in a Beta/Alpha8 Barrel Enzyme -Substrate-Bound Structures Shed New Light on the Catalytic Cycle of Hisa
J.Biol.Chem., 290, 2015
|
|
