8VUQ
 
 | |
8VUR
 
 | | Human GluN1-2A with IgG 003-102 WT conformation | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5ITI
 
 | | A cynobacterial PP2C (tPphA) structure | | Descriptor: | CALCIUM ION, Protein serin-threonin phosphatase | | Authors: | Su, J.Y. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of a Cyanobacterial PP2C Phosphatase Reveals Insights into Catalytic Mechanism and Substrate Recognition
Catalysts, 6, 2016
|
|
7LXE
 
 | |
7LSV
 
 | |
8VD6
 
 | |
8VUU
 
 | | Human GluN1-2B with Fab 007-168 | | Descriptor: | 007-168 Heavy, 007-168 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5VGT
 
 | | X-ray structure of bacteriophage Sf6 tail adaptor protein gp7 | | Descriptor: | CALCIUM ION, Gene 7 protein, MAGNESIUM ION | | Authors: | Tang, L, Liang, L, Zhao, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8VUV
 
 | |
8VLL
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) M100W mutant | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, PHOSPHATE ION, ... | | Authors: | Picard, M.-E, Grenier, J, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
8VS8
 
 | | Crystal structure of ADI-19425 Fab in complex with anti-idiotypic 1D3 Fab | | Descriptor: | 1D3 Heavy Chain, 1D3 Light Chain, ADI-19425 Heavy Chain, ... | | Authors: | Kher, G, Homad, L.J, McGuire, A.T, Pancera, M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A novel RSV vaccine candidate derived from anti-idiotypic antibodies targets putative neutralizing B-cells in bulk PBMCs
To Be Published
|
|
7LK4
 
 | |
6VQ5
 
 | |
8VUY
 
 | | Rat GluN1-2B with Fab 003-102 | | Descriptor: | 003-102 Heavy, 003-102 Light, Glutamate receptor ionotropic, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional mechanisms of anti-NMDAR autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6VQL
 
 | |
7LW1
 
 | | Human phosphofructokinase-1 liver type bound to activator NA-11 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B. | | Deposit date: | 2021-02-27 | | Release date: | 2022-01-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Selective activation of PFKL suppresses the phagocytic oxidative burst.
Cell, 184, 2021
|
|
8VXB
 
 | | Structure of ANT(6)-Ib from Campylobacter fetus subsp fetus complexed with hydrated streptomycin | | Descriptor: | PHOSPHATE ION, Streptomycin aminoglycoside adenylyltransferase ant(6)-Ib, [(2~{S},3~{S},4~{S},5~{R},6~{S})-2-[(2~{R},3~{R},4~{R},5~{S})-2-[(1~{R},2~{S},3~{R},4~{R},5~{S},6~{R})-2,4-bis[[azaniumylidene(azanyl)methyl]amino]-3,5,6-tris(oxidanyl)cyclohexyl]oxy-4-[bis(oxidanyl)methyl]-5-methyl-4-oxidanyl-oxolan-3-yl]oxy-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]-methyl-azanium | | Authors: | Nalam, P, Cook, P, Smith, B. | | Deposit date: | 2024-02-04 | | Release date: | 2024-09-18 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of Aminoglycoside Nucleotidyltransferase(6)-Ib From Campylobacter fetus subsp. fetus.
Proteins, 93, 2025
|
|
8VPG
 
 | |
6VS0
 
 | | protein B | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
5VD7
 
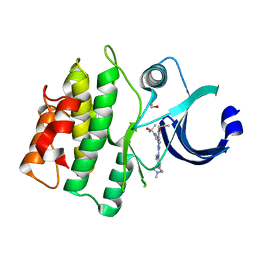 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH RAC-IV-098, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
8VT9
 
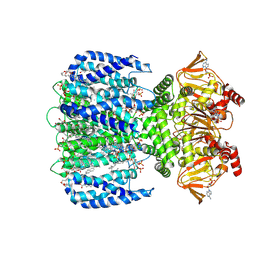 | | WT SthK in the presence of PIP2 and cAMP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, ... | | Authors: | Schmidpeter, P.A.M, Thon, O, Nimigean, C.M. | | Deposit date: | 2024-01-26 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | PIP2 inhibits pore opening of the cyclic nucleotide-gated channel SthK.
Nat Commun, 15, 2024
|
|
7LJG
 
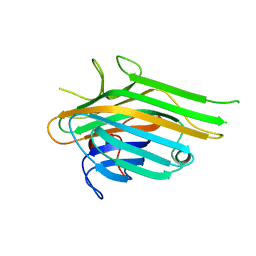 | | Crystal Structure of Lectin from Dioclea altissima | | Descriptor: | Lectin, MANGANESE (II) ION | | Authors: | Vieira-Neto, A.E, Pereira, H.M, Sousa, F.D, Goncalves, N.G.G, Vieira, N.C.G, Monteiro-Moreira, A.C.O, Moereira, R.A. | | Deposit date: | 2021-01-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Lectin from Dioclea altissima
To Be Published
|
|
8VTB
 
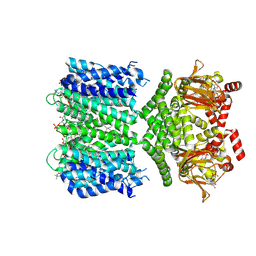 | | SthK R120A R124A in the presence of PIP2 and cAMP | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, ... | | Authors: | Schmidpeter, P.A.M, Thon, O, Nimigean, C.M. | | Deposit date: | 2024-01-26 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | PIP2 inhibits pore opening of the cyclic nucleotide-gated channel SthK.
Nat Commun, 15, 2024
|
|
7LK7
 
 | |
5I8U
 
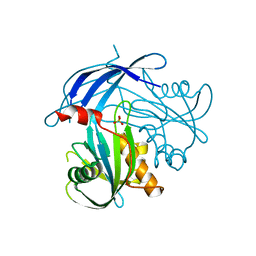 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
