9C8V
 
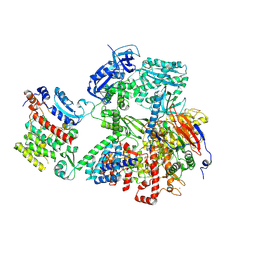 | | Human DNA polymerase alpha/primase - CHAPSO (4 mM) | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Small LEA proteins mitigate air-water interface damage to fragile cryo-EM samples during plunge freezing.
Nat Commun, 15, 2024
|
|
7X5L
 
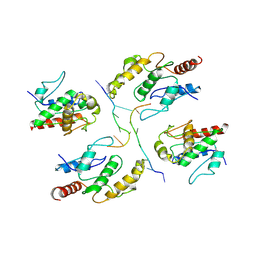 | | Tir-dsDNA complex, the initial binding state | | Descriptor: | DNA (5'-D(*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*AP*TP*TP*AP*A)-3'), Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2022-03-04 | | Release date: | 2022-06-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
1TPM
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
3S85
 
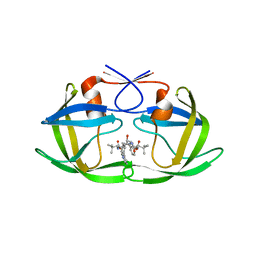 | | Discovery of New HIV Protease Inhibitors with Potential for Convenient Dosing and Reduced Side Effects: A-790742 and A-792611. | | Descriptor: | Protease/reverse transcriptase, methyl N-[(2S)-1-[[(2S,3S,5S)-5-[[(2S)-2-(methoxycarbonylamino)-3,3-dimethyl-butanoyl]amino]-3-oxidanyl-6-phenyl-1-(4-pyridin-3-ylphenyl)hexan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | DeGoey, D.A, Flosi, W.J, Grampovnik, D.J, Flentge, C.A. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus protease inhibitors (A-792611 and A-790742) with potential for convenient dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
1U15
 
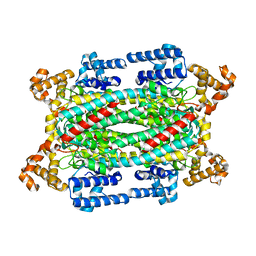 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1TPN
 
 | | SOLUTION STRUCTURE OF THE FIBRIN BINDING FINGER DOMAIN OF TISSUE-TYPE PLASMINOGEN ACTIVATOR DETERMINED BY 1H NUCLEAR MAGNETIC RESONANCE | | Descriptor: | TISSUE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Downing, A.K, Driscoll, P.C, Harvey, T.S, Dudgeon, T.J, Smith, B.O, Baron, M, Campbell, I.D. | | Deposit date: | 1993-05-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibrin binding finger domain of tissue-type plasminogen activator determined by 1H nuclear magnetic resonance.
J.Mol.Biol., 225, 1992
|
|
1TTL
 
 | | Omega-conotoxin GVIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-23 | | Release date: | 2004-07-13 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
1U16
 
 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
6TL2
 
 | |
3SH3
 
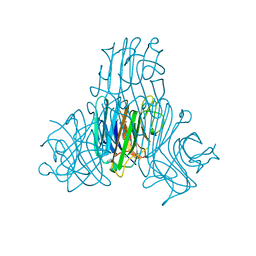 | | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii STANDL | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rangel, T.B.A, Rocha, B.A.M, Bezerra, G.A, Bezerra, M.J.B, Nascimento, K.S, Nagano, C.S, Sampaio, A.H, Assreuy, A.M.S, Gruber, K, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Biochimie, 94, 2012
|
|
1U8T
 
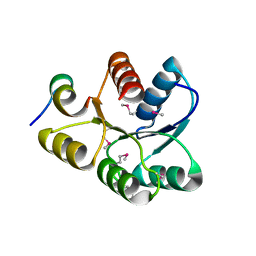 | | Crystal structure of CheY D13K Y106W alone and in complex with a FliM peptide | | Descriptor: | Chemotaxis protein cheY, Flagellar motor switch protein fliM, SULFATE ION | | Authors: | Dyer, C.M, Quillin, M.L, Campos, A, Lu, J, McEvoy, M.M, Hausrath, A.C, Westbrook, E.M, Matsumura, P, Matthews, B.W, Dahlquist, F.W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Constitutively Active Double Mutant CheY(D13K Y106W) Alone and in Complex with a FliM Peptide
J.Mol.Biol., 342, 2004
|
|
1TR6
 
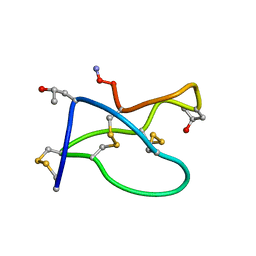 | | NMR solution structure of omega-conotoxin [K10]GVIA, a cyclic cysteine knot peptide | | Descriptor: | Omega-conotoxin GVIA | | Authors: | Mould, J, Yasuda, T, Schroeder, C.I, Beedle, A.M, Doering, C.J, Zamponi, G.W, Adams, D.J, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-13 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | The alpha2delta auxiliary subunit reduces affinity of omega-conotoxins for recombinant N-type (Cav2.2) calcium channels
J.Biol.Chem., 279, 2004
|
|
6TNF
 
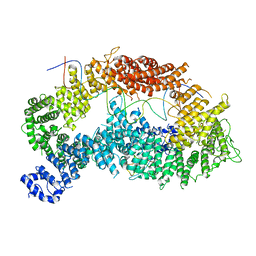 | | Structure of monoubiquitinated FANCD2 in complex with FANCI and DNA | | Descriptor: | DNA (33-MER), FANCD2, Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1U08
 
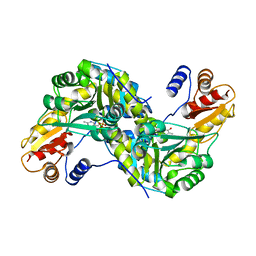 | | Crystal Structure and Reactivity of YbdL from Escherichia coli Identify a Methionine Aminotransferase Function. | | Descriptor: | Hypothetical aminotransferase ybdL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dolzan, M, Johansson, K, Roig-Zamboni, V, Campanacci, V, Tegoni, M, Schneider, G, Cambillau, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and reactivity of YbdL from Escherichia coli identify a methionine aminotransferase function
FEBS Lett., 571, 2004
|
|
1U68
 
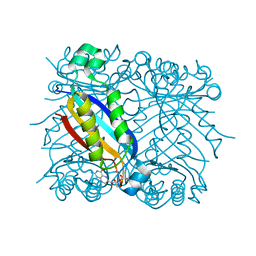 | | DHNA 7,8 DIHYDRONEOPTERIN COMPLEX | | Descriptor: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent inhibitors of dihydroneopterin aldolase using CrystaLEAD high-throughput X-ray crystallographic screening and structure-directed lead optimization.
J.MED.CHEM., 47, 2004
|
|
7MLK
 
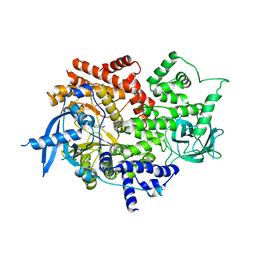 | | Crystal structure of human PI3Ka (p110a subunit) with MMV085400 bound to the active site determined at 2.9 angstroms resolution | | Descriptor: | 4-[6-(3,4,5-trimethoxyanilino)pyrazin-2-yl]benzamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Krake, S.H, Martinez, P.D.G, Poggi, M.L, Ferreira, M.S, Aguiar, A.C.C, Souza, G.E, Wenlock, M, Jones, B, Steinbrecher, T, Day, T, McPhail, J, Burke, J, Yeo, T, Mok, S, Uhlemann, A.C, Fidock, D.A, Chen, P, Grodsky, N, Deng, Y.L, Guido, R.V.C, Campbell, S.F, Willis, P.A, Dias, L.C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazines as potent PI4K inhibitors with antimalarial activity
To Be Published
|
|
1KPS
 
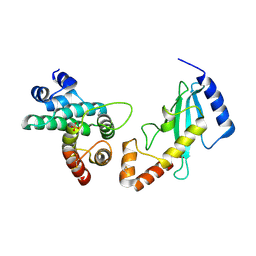 | | Structural Basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin conjugating enzyme Ubc9 and RanGAP1 | | Descriptor: | Ran-GTPase activating protein 1, SULFATE ION, Ubiquitin-like protein SUMO-1 conjugating enzyme | | Authors: | Bernier-Villamor, V, Sampson, D.A, Matunis, M.J, Lima, C.D. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1.
Cell(Cambridge,Mass.), 108, 2002
|
|
2VHH
 
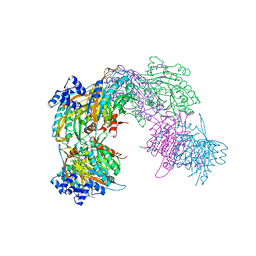 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
4U2W
 
 | | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin | | Descriptor: | 1,2-ETHANEDIOL, Bowman-Birk trypsin inhibitor, CALCIUM ION, ... | | Authors: | Grudnik, P, Golik, P, Malicki, S, Debowski, D, Legowska, A, Rolka, K, Dubin, G. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution crystal structure of HV-BBI protease inhibitor from amphibian skin in complex with bovine trypsin.
Proteins, 83, 2015
|
|
2VA5
 
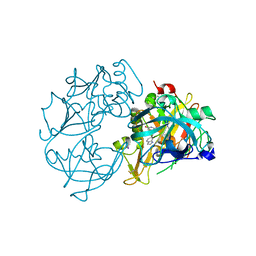 | | X-ray crystal structure of beta secretase complexed with compound 8c | | Descriptor: | 2-amino-6-[2-(1H-indol-6-yl)ethyl]pyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
2VER
 
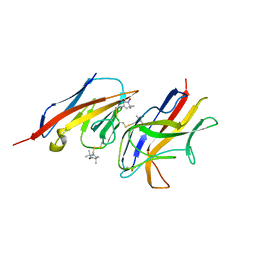 | | Structural model for the complex between the Dr adhesins and carcinoembryonic antigen (CEA) | | Descriptor: | AFIMBRIAL ADHESIN AFA-III, ARCINOEMBRYONIC ANTIGEN-RELATED CELL ADHESION MOLECULE 5, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Korotkova, N, Yang, Y, Le Trong, I, Cota, E, Demeler, B, Marchant, J, Thomas, W.E, Stenkamp, R.E, Moseley, S.L, Matthews, S. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Binding of Dr Adhesins of Escherichia Coli to Carcinoembryonic Antigen Triggers Receptor Dissociation.
Mol.Microbiol., 67, 2008
|
|
2VH6
 
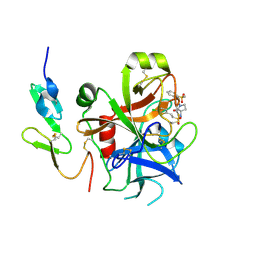 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with biaryl P4 motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-{(3S)-1-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-2-oxopyrrolidin-3-yl}ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Chung, C.W, Convery, M.A, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A.N, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7PLM
 
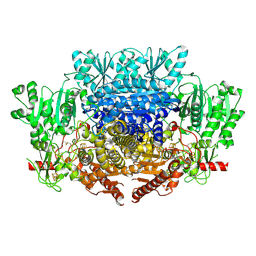 | | CryoEM reconstruction of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic conditions | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Cherrier, M.V, Vernede, X, Fenel, D, Martin, L, Arragain, B, Neumann, E, Fontecilla Camps, J.C, Schoehn, G, Nicolet, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Oxygen-Sensitive Metalloprotein Structure Determination by Cryo-Electron Microscopy.
Biomolecules, 12, 2022
|
|
2VGH
 
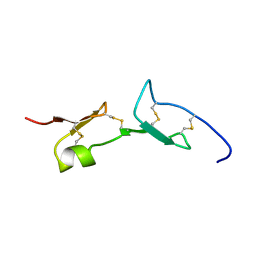 | | HEPARIN-BINDING DOMAIN FROM VASCULAR ENDOTHELIAL GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR-165 | | Authors: | Fairbrother, W.J, Champe, M.A, Christinger, H.W, Keyt, B.A, Starovasnik, M.A. | | Deposit date: | 1997-12-17 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heparin-binding domain of vascular endothelial growth factor.
Structure, 6, 1998
|
|
2VA7
 
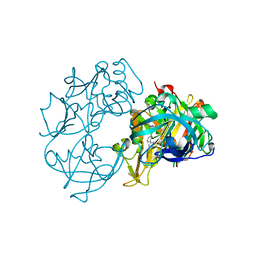 | | X-ray crystal structure of beta secretase complexed with compound 27 | | Descriptor: | (6R)-2-amino-6-[2-(3'-methoxybiphenyl-3-yl)ethyl]-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, BETA-SECRETASE 1 ., IODIDE ION | | Authors: | Edwards, P.D, Albert, J.S, Sylvester, M, Aharony, D, Andisik, D, Callaghan, O, Campbell, J.B, Carr, R.A, Chessari, G, Congreve, M, Frederickson, M, Folmer, R.H.A, Geschwindner, S, Koether, G, Kolmodin, K, Krumrine, J, Mauger, R.C, Murray, C.W, Olsson, L.L, Patel, S, Spear, N, Tian, G. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-Based Lead Generation to the Discovery of Novel, Cyclic Amidine Beta-Secretase Inhibitors with Nanomolar Potency, Cellular Activity, and High Ligand Efficiency.
J.Med.Chem., 50, 2007
|
|
