4LH6
 
 | | Crystal structure of a LigA inhibitor | | Descriptor: | 4-amino-2-bromothieno[3,2-c]pyridine-7-carboxamide, ACETATE ION, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, ... | | Authors: | Benenato, K, Wang, H, Mcguire, H.M, Davis, H, Gao, N, Prince, D.B, Jahic, H, Stokes, S.S, Boriack-Sjodin, P.A. | | Deposit date: | 2013-06-30 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification through structure-based methods of a bacterial NAD(+)-dependent DNA ligase inhibitor that avoids known resistance mutations.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7RPW
 
 | |
7RPO
 
 | |
7RPX
 
 | |
7UFP
 
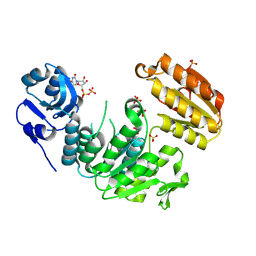 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | Mur ligase middle domain protein, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-03-23 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
4E51
 
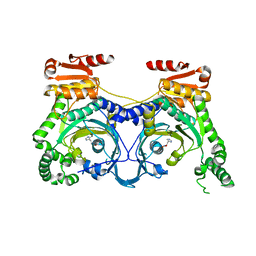 | |
8RHZ
 
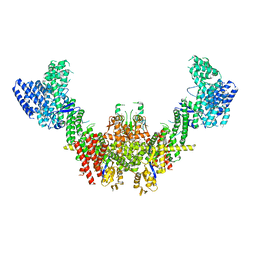 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated conformation - symmetry expanded unneddylated dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2023-12-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4LJP
 
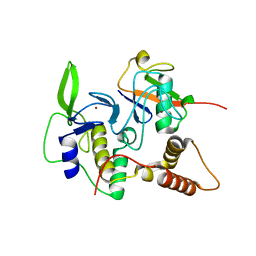 | | Structure of an active ligase (HOIP-H889A)/ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-C, ZINC ION | | Authors: | Rana, R.R, Stieglitz, B, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
8ST7
 
 | | Structure of E3 ligase VsHECT bound to ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase SopA-like catalytic domain-containing protein, Ubiquitin, prop-2-en-1-amine | | Authors: | Franklin, T.G, Pruneda, J.N. | | Deposit date: | 2023-05-09 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Bacterial ligases reveal fundamental principles of polyubiquitin specificity.
Mol.Cell, 83, 2023
|
|
8ST9
 
 | |
9FZX
 
 | | Rhizobium phage ligase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, DNA, ... | | Authors: | Rothweiler, U, Williamson, A. | | Deposit date: | 2024-07-06 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal structure of ATP-dependent DNA ligase from Rhizobium phage vB_RleM_P10VF.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
8Q7E
 
 | |
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3N84
 
 | |
4LJQ
 
 | | Crystal structure of the catalytic core of E3 ligase HOIP | | Descriptor: | E3 ubiquitin-protein ligase RNF31, ZINC ION | | Authors: | Stieglitz, B, Rana, R.R, Koliopoulos, M.G, Morris-Davies, A.C, Christodoulou, E, Howell, S, Brown, N.R, Rittinger, K. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP.
Nature, 503, 2013
|
|
8ST8
 
 | |
8D4X
 
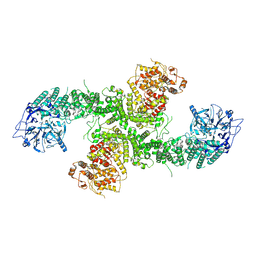 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
8AK4
 
 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
9DC3
 
 | | AAV8 in complex with the AAVX affinity ligand | | Descriptor: | AAVX affinity ligand, Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2024-08-25 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structural characterization and epitope mapping of the AAVX affinity purification ligand.
Mol Ther Methods Clin Dev, 32, 2024
|
|
4L9P
 
 | | Crystal structure of Aspergillus fumigatus protein farnesyltransferase complexed with the FII analog, FPT-II, and the KCVVM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CaaX farnesyltransferase alpha subunit Ram2, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-06-18 | | Release date: | 2014-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
4LNG
 
 | | Aspergillus fumigatus protein farnesyltransferase complex with farnesyldiphosphate and tipifarnib | | Descriptor: | 1,2-ETHANEDIOL, 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, CaaX farnesyltransferase alpha subunit Ram2, ... | | Authors: | Mabanglo, M.F, Hast, M.A, Beese, L.S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design.
Protein Sci., 23, 2014
|
|
4UYM
 
 | |
4D05
 
 | | Structure and activity of a minimal-type ATP-dependent DNA ligase from a psychrotolerant bacterium | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-DEPENDENT DNA LIGASE, MAGNESIUM ION, ... | | Authors: | Williamson, A, Rothweiler, U, Leiros, H.-K.S. | | Deposit date: | 2014-04-24 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme-Adenylate Structure of a Bacterial ATP-Dependent DNA Ligase with a Minimized DNA-Binding Surface
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3PN1
 
 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | Authors: | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
6SAL
 
 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Doveston, R.G, Brunsveld, L. | | Deposit date: | 2019-07-17 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Ligand-Based Design of Allosteric Retinoic Acid Receptor-Related Orphan Receptor gamma t (ROR gamma t) Inverse Agonists.
J.Med.Chem., 63, 2020
|
|
