6FL0
 
 | | Crystal structure of the membrane attack complex assembly inhibitor BGA71 from Lyme disease agent Borreliella bavariensis | | Descriptor: | membrane attack complex assembly inhibitor BGA71 | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2018-01-25 | | Release date: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the membrane attack complex assembly inhibitor BGA71 from the Lyme disease agent Borrelia bavariensis.
Sci Rep, 8, 2018
|
|
8GCA
 
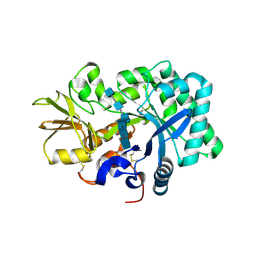 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N',N''-triacetylchitotriose at pH 4.74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
7OXO
 
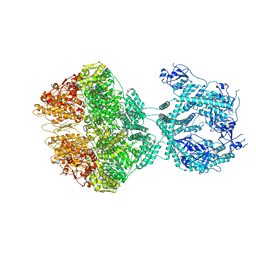 | | human LonP1, R-state, incubated in AMPPCP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Abrahams, J.P, Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T. | | Deposit date: | 2021-06-22 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
8T5A
 
 | |
4JYW
 
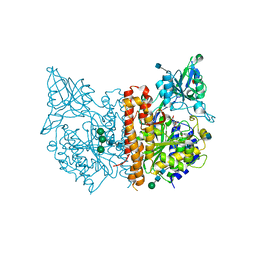 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with CTT1057 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C. | | Deposit date: | 2013-04-01 | | Release date: | 2014-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A high affinity 18F-labeled phosphoramidate peptidomimetic inhibitor as a PSMA-targeted PET imaging agent for prostate cancer
To be Published
|
|
7P3B
 
 | | Human RNA ligase RTCB in complex with GMP and Co(II) | | Descriptor: | ACETATE ION, COBALT (II) ION, FORMIC ACID, ... | | Authors: | Kroupova, A, Ackle, F, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
4K8M
 
 | | High resolution structure of M.tb NRDH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Phulera, S, Mande, S.C. | | Deposit date: | 2013-04-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | The crystal structure of Mycobacterium tuberculosis NrdH at 0.87 angstrom suggests a possible mode of its activity.
Biochemistry, 52, 2013
|
|
8GAI
 
 | |
7OSM
 
 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
6G3K
 
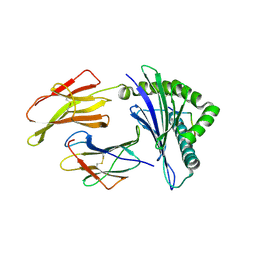 | | MHC A02 Allele presenting MTSAIGILPV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2018-03-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide Super-Agonist Enhances T-Cell Responses to Melanoma.
Front Immunol, 10, 2019
|
|
7OT7
 
 | |
7P6X
 
 | | Cryo-Em structure of the hexameric RUVBL1-RUVBL2 in complex with ZNHIT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2021-07-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of RUVBL1-RUVBL2-ZNHIT2, a complex that interacts with pre-mRNA-processing-splicing factor 8.
Nucleic Acids Res., 50, 2022
|
|
4K26
 
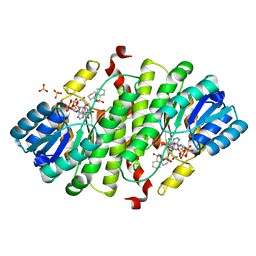 | | 4,4-Dioxo-5,6-dihydro-[1,4,3]oxathiazines, a novel class of 11 -HSD1 inhibitors for the treatment of diabetes | | Descriptor: | (4aS,8aR)-N-cyclohexyl-4a,5,6,7,8,8a-hexahydro-4,1,2-benzoxathiazin-3-amine 1,1-dioxide, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Loenze, P, Schimanski-Breves, S, Vonderheyden, C, Engel, C.K. | | Deposit date: | 2013-04-08 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 1,1-Dioxo-5,6-dihydro-[4,1,2]oxathiazines, a novel class of 11-HSD1 inhibitors for the treatment of diabetes.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8G9Z
 
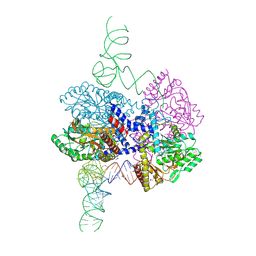 | | High-resolution crystal structure of the human selenomethionine-derived SepSecS-tRNASec complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, O-phosphoseryl-tRNA(Sec) selenium transferase, ... | | Authors: | Puppala, A, Simonovic, M, Castillo Suchkou, J. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-05 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for the tRNA-dependent activation of the terminal complex of selenocysteine synthesis in humans.
Nucleic Acids Res., 51, 2023
|
|
4K20
 
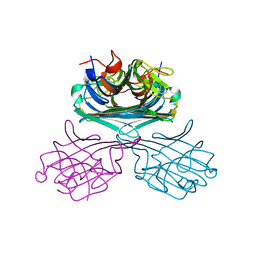 | | Crystal structure of Canavalia boliviana lectin | | Descriptor: | CALCIUM ION, Canavalia boliviana lectin, MANGANESE (II) ION | | Authors: | Viertlmayr, R, Bezerra, G.A, Moura, T.R, Delatorre, P, Rocha, B.A.M, Cavada, B.S, Gruber, K. | | Deposit date: | 2013-04-07 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Studies of an Anti-Inflammatory Lectin from Canavalia boliviana Seeds in Complex with Dimannosides.
Plos One, 9, 2014
|
|
6GB6
 
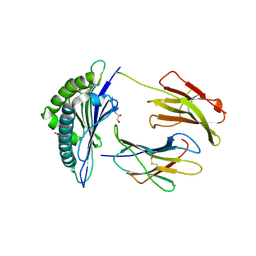 | | Structure of H-2Kb with dipeptide GL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCINE, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7P9D
 
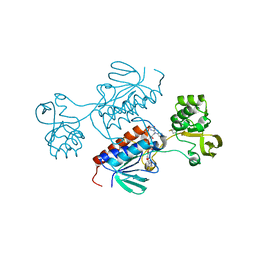 | | Crystal structure of Chlamydomonas reinhardtii NADPH Dependent Thioredoxin Reductase 1 domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Singh, R.K, Marchetti, G.M, Hippler, M, Kuemmel, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis revealed a novel conformation of the NTRC reductase domain from Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2021
|
|
6GBF
 
 | |
8TD4
 
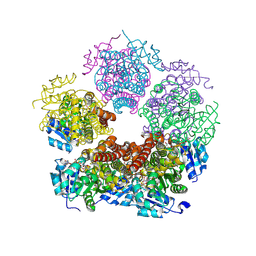 | | Structure of PYCR1 complexed with NADH and 1,3-Dithiolane-2-carboxylic acid | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
7OCX
 
 | |
6FXP
 
 | | Crystal structure of S. aureus glucosaminidase B | | Descriptor: | CHLORIDE ION, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Pintar, S, Turk, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Domain sliding of two Staphylococcus aureus N-acetylglucosaminidases enables their substrate-binding prior to its catalysis.
Commun Biol, 3, 2020
|
|
4JZW
 
 | |
6G5O
 
 | | The structure of a carbohydrate active P450 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Robb, C.S, Hehemann, J.H. | | Deposit date: | 2018-03-29 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specificity and mechanism of carbohydrate demethylation by cytochrome P450 monooxygenases.
Biochem. J., 475, 2018
|
|
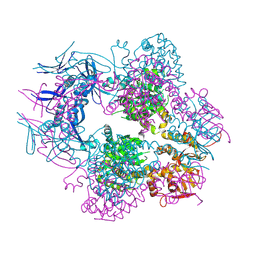
8TCH
 
 | |
6G5Z
 
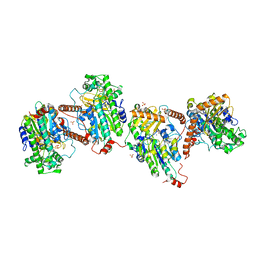 | |
